
Schistosoma margrebowiei
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Spiralia; Lophotrochozoa; Platyhelminthes; Trematoda; Digenea; Strigeidida; Schistosomatoidea; Schistosomatidae; Schistosoma
Average proteome isoelectric point is 7.03
Get precalculated fractions of proteins
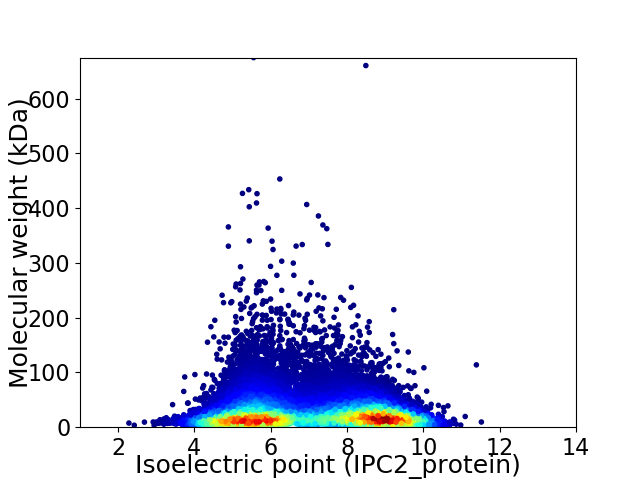
Virtual 2D-PAGE plot for 25527 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A183LRR1|A0A183LRR1_9TREM Uncharacterized protein OS=Schistosoma margrebowiei OX=48269 GN=SMRZ_LOCUS6486 PE=4 SV=1
FF1 pKa = 7.13SAGISEE7 pKa = 4.57IPVSTTANTSHH18 pKa = 7.43LDD20 pKa = 3.61YY21 pKa = 11.31SQLDD25 pKa = 3.91LNDD28 pKa = 4.35DD29 pKa = 4.25LVAKK33 pKa = 10.14DD34 pKa = 4.18LQLTDD39 pKa = 4.6SDD41 pKa = 5.57DD42 pKa = 4.62DD43 pKa = 5.6DD44 pKa = 6.39GDD46 pKa = 4.07SSLTGQGGEE55 pKa = 4.23LCTDD59 pKa = 3.51EE60 pKa = 5.55NDD62 pKa = 3.72ATVMSYY68 pKa = 11.05NIDD71 pKa = 3.53QDD73 pKa = 4.18NDD75 pKa = 3.72DD76 pKa = 5.89DD77 pKa = 5.46DD78 pKa = 6.14DD79 pKa = 4.23QMHH82 pKa = 7.24RR83 pKa = 11.84NTILQYY89 pKa = 10.09TEE91 pKa = 3.69YY92 pKa = 9.75HH93 pKa = 5.65QNEE96 pKa = 4.66SEE98 pKa = 4.41MISHH102 pKa = 7.23NDD104 pKa = 3.37NSFTTYY110 pKa = 10.25NVQHH114 pKa = 6.56RR115 pKa = 11.84QRR117 pKa = 11.84LDD119 pKa = 3.34HH120 pKa = 7.74DD121 pKa = 4.62DD122 pKa = 3.85SNNNNNHH129 pKa = 5.79NNSHH133 pKa = 5.85PTFFISDD140 pKa = 4.42DD141 pKa = 4.31DD142 pKa = 6.05DD143 pKa = 6.0DD144 pKa = 6.07DD145 pKa = 3.9NN146 pKa = 5.8
FF1 pKa = 7.13SAGISEE7 pKa = 4.57IPVSTTANTSHH18 pKa = 7.43LDD20 pKa = 3.61YY21 pKa = 11.31SQLDD25 pKa = 3.91LNDD28 pKa = 4.35DD29 pKa = 4.25LVAKK33 pKa = 10.14DD34 pKa = 4.18LQLTDD39 pKa = 4.6SDD41 pKa = 5.57DD42 pKa = 4.62DD43 pKa = 5.6DD44 pKa = 6.39GDD46 pKa = 4.07SSLTGQGGEE55 pKa = 4.23LCTDD59 pKa = 3.51EE60 pKa = 5.55NDD62 pKa = 3.72ATVMSYY68 pKa = 11.05NIDD71 pKa = 3.53QDD73 pKa = 4.18NDD75 pKa = 3.72DD76 pKa = 5.89DD77 pKa = 5.46DD78 pKa = 6.14DD79 pKa = 4.23QMHH82 pKa = 7.24RR83 pKa = 11.84NTILQYY89 pKa = 10.09TEE91 pKa = 3.69YY92 pKa = 9.75HH93 pKa = 5.65QNEE96 pKa = 4.66SEE98 pKa = 4.41MISHH102 pKa = 7.23NDD104 pKa = 3.37NSFTTYY110 pKa = 10.25NVQHH114 pKa = 6.56RR115 pKa = 11.84QRR117 pKa = 11.84LDD119 pKa = 3.34HH120 pKa = 7.74DD121 pKa = 4.62DD122 pKa = 3.85SNNNNNHH129 pKa = 5.79NNSHH133 pKa = 5.85PTFFISDD140 pKa = 4.42DD141 pKa = 4.31DD142 pKa = 6.05DD143 pKa = 6.0DD144 pKa = 6.07DD145 pKa = 3.9NN146 pKa = 5.8
Molecular weight: 16.54 kDa
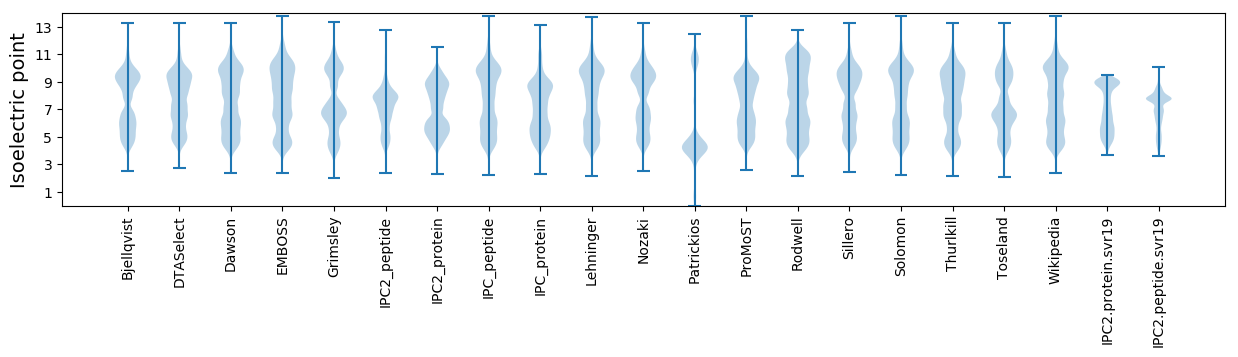
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3P8CR17|A0A3P8CR17_9TREM Uncharacterized protein OS=Schistosoma margrebowiei OX=48269 GN=SMRZ_LOCUS9523 PE=4 SV=1
MM1 pKa = 7.73LILLRR6 pKa = 11.84LILLLLLQRR15 pKa = 11.84LLRR18 pKa = 11.84LPRR21 pKa = 11.84LLLLQRR27 pKa = 11.84LLRR30 pKa = 11.84LLPRR34 pKa = 11.84LILLVLLLLLLLLLLLRR51 pKa = 11.84LPLLRR56 pKa = 11.84LLLLLLRR63 pKa = 11.84LLLLLLLLLLLIRR76 pKa = 11.84LLLLLYY82 pKa = 10.7LLLRR86 pKa = 11.84RR87 pKa = 11.84RR88 pKa = 11.84LLLLRR93 pKa = 11.84LLRR96 pKa = 11.84LLLLLLLLLLLRR108 pKa = 11.84LLLLRR113 pKa = 11.84ILLILLLLLLLLLLLLLLLRR133 pKa = 11.84LLLLLRR139 pKa = 11.84LLRR142 pKa = 11.84LLLLLLLRR150 pKa = 11.84LLLLLQLLLLLRR162 pKa = 11.84LLLRR166 pKa = 11.84LLLLLLLLLLILLLLLLLLLLIIIIILLLLLLLLLILLLLRR207 pKa = 11.84LIRR210 pKa = 11.84LQLLLLLLLILYY222 pKa = 9.87YY223 pKa = 10.46YY224 pKa = 10.52DD225 pKa = 3.32YY226 pKa = 11.11YY227 pKa = 11.68YY228 pKa = 10.49PLLLLQLLLLLLLPLLRR245 pKa = 11.84LLLLPRR251 pKa = 11.84LLRR254 pKa = 11.84LQRR257 pKa = 11.84LQLLLLRR264 pKa = 11.84LLLLLLLLLLLPLRR278 pKa = 11.84LRR280 pKa = 11.84LLLLLLLLLLLILLLLLLLLLLLLLLILLLLRR312 pKa = 11.84LLLLLVLLLLLLLLLRR328 pKa = 11.84LLLLRR333 pKa = 11.84LLLRR337 pKa = 11.84VLLLLLLLLRR347 pKa = 11.84LLLLLLLLLLLLLLLIIIIILLLLLLLLILLLLRR381 pKa = 11.84LIRR384 pKa = 11.84LQPLLLLLLLLLLLLLLLLLLRR406 pKa = 11.84LLLLRR411 pKa = 11.84LLLRR415 pKa = 11.84LLLLLRR421 pKa = 11.84LLRR424 pKa = 11.84ILLLLLLLLLLLLLLLILQRR444 pKa = 11.84VVPLRR449 pKa = 11.84LLLLLLLLIIIILIIIIIILLLLLLPLSRR478 pKa = 11.84LLRR481 pKa = 11.84LQRR484 pKa = 11.84LILLLRR490 pKa = 11.84LLLLLLLLLLLLQRR504 pKa = 11.84LLRR507 pKa = 11.84LLLLQRR513 pKa = 11.84ILRR516 pKa = 11.84LQRR519 pKa = 11.84LLLILLLLLPLLRR532 pKa = 11.84LLLLLLLLLLLIRR545 pKa = 11.84LLLLRR550 pKa = 11.84LLLLLRR556 pKa = 11.84LLRR559 pKa = 11.84LLLLLRR565 pKa = 11.84LLLLLLLLLLLLLRR579 pKa = 11.84LLQRR583 pKa = 11.84LLRR586 pKa = 11.84LLLILLLLLLLRR598 pKa = 11.84QLLILLLLLILLLLLLLLLLLLLRR622 pKa = 11.84LLLLRR627 pKa = 11.84LLLRR631 pKa = 11.84LLRR634 pKa = 11.84ILLLLLLLLLLLLLRR649 pKa = 11.84LLLLLLLLIIIIIIILLPLLLLLLLLLLLILQRR682 pKa = 11.84LVPLRR687 pKa = 11.84LLLLLLLIIIIIIIIIIIKK706 pKa = 10.11IIIKK710 pKa = 10.25IILLLLLLLLLLLLLRR726 pKa = 11.84LILLLRR732 pKa = 11.84LLPLLLLLLLLLLLLLLRR750 pKa = 11.84ILPLPRR756 pKa = 11.84LLRR759 pKa = 11.84LQCLIILLRR768 pKa = 11.84LLLLLLLLLLLLQRR782 pKa = 11.84LPRR785 pKa = 11.84LLLLQRR791 pKa = 11.84ILRR794 pKa = 11.84LQRR797 pKa = 11.84LLLLLLLLLPLLRR810 pKa = 11.84LLLLLLLLLIIIIILLPLQHH830 pKa = 7.1LLLLLLLLLLLILLLLLLLLLLLILLLLLLLLLLLLLLLRR870 pKa = 11.84LLLLLLLLLLLLLLLLLLLLPLLRR894 pKa = 11.84LLLLLLLLLLRR905 pKa = 11.84LLQRR909 pKa = 11.84LLRR912 pKa = 11.84LLVLLLLLILILLLLLLLLLLRR934 pKa = 11.84LLLLLRR940 pKa = 11.84LLPLLLLLLLLLLLLRR956 pKa = 11.84LLPLPRR962 pKa = 11.84LLRR965 pKa = 11.84LQQ967 pKa = 3.56
MM1 pKa = 7.73LILLRR6 pKa = 11.84LILLLLLQRR15 pKa = 11.84LLRR18 pKa = 11.84LPRR21 pKa = 11.84LLLLQRR27 pKa = 11.84LLRR30 pKa = 11.84LLPRR34 pKa = 11.84LILLVLLLLLLLLLLLRR51 pKa = 11.84LPLLRR56 pKa = 11.84LLLLLLRR63 pKa = 11.84LLLLLLLLLLLIRR76 pKa = 11.84LLLLLYY82 pKa = 10.7LLLRR86 pKa = 11.84RR87 pKa = 11.84RR88 pKa = 11.84LLLLRR93 pKa = 11.84LLRR96 pKa = 11.84LLLLLLLLLLLRR108 pKa = 11.84LLLLRR113 pKa = 11.84ILLILLLLLLLLLLLLLLLRR133 pKa = 11.84LLLLLRR139 pKa = 11.84LLRR142 pKa = 11.84LLLLLLLRR150 pKa = 11.84LLLLLQLLLLLRR162 pKa = 11.84LLLRR166 pKa = 11.84LLLLLLLLLLILLLLLLLLLLIIIIILLLLLLLLLILLLLRR207 pKa = 11.84LIRR210 pKa = 11.84LQLLLLLLLILYY222 pKa = 9.87YY223 pKa = 10.46YY224 pKa = 10.52DD225 pKa = 3.32YY226 pKa = 11.11YY227 pKa = 11.68YY228 pKa = 10.49PLLLLQLLLLLLLPLLRR245 pKa = 11.84LLLLPRR251 pKa = 11.84LLRR254 pKa = 11.84LQRR257 pKa = 11.84LQLLLLRR264 pKa = 11.84LLLLLLLLLLLPLRR278 pKa = 11.84LRR280 pKa = 11.84LLLLLLLLLLLILLLLLLLLLLLLLLILLLLRR312 pKa = 11.84LLLLLVLLLLLLLLLRR328 pKa = 11.84LLLLRR333 pKa = 11.84LLLRR337 pKa = 11.84VLLLLLLLLRR347 pKa = 11.84LLLLLLLLLLLLLLLIIIIILLLLLLLLILLLLRR381 pKa = 11.84LIRR384 pKa = 11.84LQPLLLLLLLLLLLLLLLLLLRR406 pKa = 11.84LLLLRR411 pKa = 11.84LLLRR415 pKa = 11.84LLLLLRR421 pKa = 11.84LLRR424 pKa = 11.84ILLLLLLLLLLLLLLLILQRR444 pKa = 11.84VVPLRR449 pKa = 11.84LLLLLLLLIIIILIIIIIILLLLLLPLSRR478 pKa = 11.84LLRR481 pKa = 11.84LQRR484 pKa = 11.84LILLLRR490 pKa = 11.84LLLLLLLLLLLLQRR504 pKa = 11.84LLRR507 pKa = 11.84LLLLQRR513 pKa = 11.84ILRR516 pKa = 11.84LQRR519 pKa = 11.84LLLILLLLLPLLRR532 pKa = 11.84LLLLLLLLLLLIRR545 pKa = 11.84LLLLRR550 pKa = 11.84LLLLLRR556 pKa = 11.84LLRR559 pKa = 11.84LLLLLRR565 pKa = 11.84LLLLLLLLLLLLLRR579 pKa = 11.84LLQRR583 pKa = 11.84LLRR586 pKa = 11.84LLLILLLLLLLRR598 pKa = 11.84QLLILLLLLILLLLLLLLLLLLLRR622 pKa = 11.84LLLLRR627 pKa = 11.84LLLRR631 pKa = 11.84LLRR634 pKa = 11.84ILLLLLLLLLLLLLRR649 pKa = 11.84LLLLLLLLIIIIIIILLPLLLLLLLLLLLILQRR682 pKa = 11.84LVPLRR687 pKa = 11.84LLLLLLLIIIIIIIIIIIKK706 pKa = 10.11IIIKK710 pKa = 10.25IILLLLLLLLLLLLLRR726 pKa = 11.84LILLLRR732 pKa = 11.84LLPLLLLLLLLLLLLLLRR750 pKa = 11.84ILPLPRR756 pKa = 11.84LLRR759 pKa = 11.84LQCLIILLRR768 pKa = 11.84LLLLLLLLLLLLQRR782 pKa = 11.84LPRR785 pKa = 11.84LLLLQRR791 pKa = 11.84ILRR794 pKa = 11.84LQRR797 pKa = 11.84LLLLLLLLLPLLRR810 pKa = 11.84LLLLLLLLLIIIIILLPLQHH830 pKa = 7.1LLLLLLLLLLLILLLLLLLLLLLILLLLLLLLLLLLLLLRR870 pKa = 11.84LLLLLLLLLLLLLLLLLLLLPLLRR894 pKa = 11.84LLLLLLLLLLRR905 pKa = 11.84LLQRR909 pKa = 11.84LLRR912 pKa = 11.84LLVLLLLLILILLLLLLLLLLRR934 pKa = 11.84LLLLLRR940 pKa = 11.84LLPLLLLLLLLLLLLRR956 pKa = 11.84LLPLPRR962 pKa = 11.84LLRR965 pKa = 11.84LQQ967 pKa = 3.56
Molecular weight: 113.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6596622 |
29 |
5872 |
258.4 |
29.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.618 ± 0.018 | 2.033 ± 0.011 |
5.219 ± 0.014 | 5.551 ± 0.019 |
3.714 ± 0.013 | 4.522 ± 0.021 |
2.901 ± 0.01 | 6.739 ± 0.019 |
6.001 ± 0.019 | 9.515 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.165 ± 0.007 | 7.219 ± 0.03 |
4.269 ± 0.017 | 4.386 ± 0.013 |
5.15 ± 0.017 | 9.651 ± 0.031 |
6.768 ± 0.027 | 5.318 ± 0.015 |
1.153 ± 0.007 | 3.103 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
