
Sanxia tombus-like virus 4
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.6
Get precalculated fractions of proteins
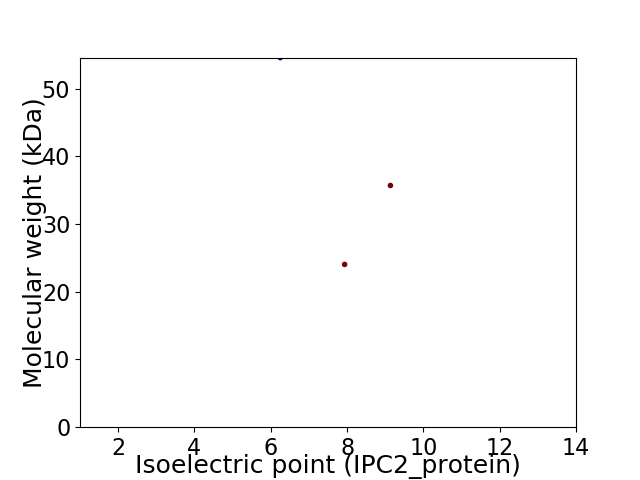
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KGK5|A0A1L3KGK5_9VIRU RNA-directed RNA polymerase OS=Sanxia tombus-like virus 4 OX=1923388 PE=4 SV=1
MM1 pKa = 7.74DD2 pKa = 4.29VKK4 pKa = 10.73PFEE7 pKa = 4.25KK8 pKa = 10.25TLDD11 pKa = 3.72NVVSGIKK18 pKa = 9.74EE19 pKa = 4.01RR20 pKa = 11.84IMYY23 pKa = 9.94IDD25 pKa = 3.96DD26 pKa = 4.02AGTRR30 pKa = 11.84RR31 pKa = 11.84PVCRR35 pKa = 11.84RR36 pKa = 11.84NAGEE40 pKa = 4.58LKK42 pKa = 10.73HH43 pKa = 6.41LTDD46 pKa = 4.62KK47 pKa = 11.3LADD50 pKa = 3.75CVPNPTRR57 pKa = 11.84MTRR60 pKa = 11.84RR61 pKa = 11.84EE62 pKa = 4.14FIASRR67 pKa = 11.84NGRR70 pKa = 11.84LKK72 pKa = 10.52KK73 pKa = 10.49VYY75 pKa = 9.82EE76 pKa = 4.4RR77 pKa = 11.84ANEE80 pKa = 4.0MLDD83 pKa = 3.82DD84 pKa = 4.07QPSSLEE90 pKa = 3.89DD91 pKa = 3.39LARR94 pKa = 11.84TSLFTKK100 pKa = 9.6WEE102 pKa = 4.14RR103 pKa = 11.84SVHH106 pKa = 5.9KK107 pKa = 9.82PGKK110 pKa = 6.56TTVPRR115 pKa = 11.84IINPRR120 pKa = 11.84SPQFNILLGRR130 pKa = 11.84YY131 pKa = 7.03LTPIEE136 pKa = 4.03HH137 pKa = 6.67QIFEE141 pKa = 4.63GLQEE145 pKa = 4.25MLEE148 pKa = 4.31SPHH151 pKa = 6.38PVIAKK156 pKa = 10.17GLTQSEE162 pKa = 4.43KK163 pKa = 10.61GQIIADD169 pKa = 3.84IVNDD173 pKa = 4.82GYY175 pKa = 11.72VVVGLDD181 pKa = 3.28ASRR184 pKa = 11.84FDD186 pKa = 3.41QCISEE191 pKa = 4.54EE192 pKa = 4.05LLKK195 pKa = 10.24MEE197 pKa = 4.75HH198 pKa = 6.26NVYY201 pKa = 10.33LKK203 pKa = 10.53CYY205 pKa = 10.1QNDD208 pKa = 3.66RR209 pKa = 11.84LLRR212 pKa = 11.84ALLKK216 pKa = 10.75CQLDD220 pKa = 3.36NSGQYY225 pKa = 9.39IGRR228 pKa = 11.84DD229 pKa = 3.15GRR231 pKa = 11.84VKK233 pKa = 10.4VRR235 pKa = 11.84YY236 pKa = 8.79GAIRR240 pKa = 11.84CSGDD244 pKa = 3.03MNTSLGNCIISVLLSVLFCDD264 pKa = 4.17EE265 pKa = 4.24NGIGDD270 pKa = 3.87FRR272 pKa = 11.84VFCDD276 pKa = 4.84GDD278 pKa = 4.02DD279 pKa = 3.74LLLAVRR285 pKa = 11.84RR286 pKa = 11.84GDD288 pKa = 3.91LNKK291 pKa = 10.47LNPLQQWYY299 pKa = 8.46LQWGLRR305 pKa = 11.84MKK307 pKa = 10.59VEE309 pKa = 4.41EE310 pKa = 4.05PAYY313 pKa = 9.33TPEE316 pKa = 4.14AVEE319 pKa = 4.84FCQSRR324 pKa = 11.84PVCIDD329 pKa = 2.59GRR331 pKa = 11.84YY332 pKa = 9.67VLIRR336 pKa = 11.84DD337 pKa = 3.25VRR339 pKa = 11.84KK340 pKa = 10.17CINVDD345 pKa = 2.98YY346 pKa = 11.18SGFVNLQDD354 pKa = 3.38RR355 pKa = 11.84DD356 pKa = 3.78YY357 pKa = 9.72MLKK360 pKa = 9.51YY361 pKa = 9.9LRR363 pKa = 11.84AVGVCGSYY371 pKa = 10.34LAAGCPVLQAWYY383 pKa = 9.67GFGVRR388 pKa = 11.84VGTTGKK394 pKa = 10.53LDD396 pKa = 3.45VLDD399 pKa = 3.98TEE401 pKa = 5.26RR402 pKa = 11.84GFTRR406 pKa = 11.84QAKK409 pKa = 9.67LEE411 pKa = 4.03QRR413 pKa = 11.84SGAVMYY419 pKa = 10.49RR420 pKa = 11.84PIDD423 pKa = 4.08DD424 pKa = 3.97GARR427 pKa = 11.84EE428 pKa = 4.18SFRR431 pKa = 11.84LAFGIGAAEE440 pKa = 3.81QLLLEE445 pKa = 4.74SIFDD449 pKa = 3.6EE450 pKa = 4.38MTLEE454 pKa = 4.27RR455 pKa = 11.84GQLDD459 pKa = 3.47SHH461 pKa = 6.13PQEE464 pKa = 3.84ITDD467 pKa = 4.17FSNTFAFLVQPQHH480 pKa = 5.65GG481 pKa = 3.65
MM1 pKa = 7.74DD2 pKa = 4.29VKK4 pKa = 10.73PFEE7 pKa = 4.25KK8 pKa = 10.25TLDD11 pKa = 3.72NVVSGIKK18 pKa = 9.74EE19 pKa = 4.01RR20 pKa = 11.84IMYY23 pKa = 9.94IDD25 pKa = 3.96DD26 pKa = 4.02AGTRR30 pKa = 11.84RR31 pKa = 11.84PVCRR35 pKa = 11.84RR36 pKa = 11.84NAGEE40 pKa = 4.58LKK42 pKa = 10.73HH43 pKa = 6.41LTDD46 pKa = 4.62KK47 pKa = 11.3LADD50 pKa = 3.75CVPNPTRR57 pKa = 11.84MTRR60 pKa = 11.84RR61 pKa = 11.84EE62 pKa = 4.14FIASRR67 pKa = 11.84NGRR70 pKa = 11.84LKK72 pKa = 10.52KK73 pKa = 10.49VYY75 pKa = 9.82EE76 pKa = 4.4RR77 pKa = 11.84ANEE80 pKa = 4.0MLDD83 pKa = 3.82DD84 pKa = 4.07QPSSLEE90 pKa = 3.89DD91 pKa = 3.39LARR94 pKa = 11.84TSLFTKK100 pKa = 9.6WEE102 pKa = 4.14RR103 pKa = 11.84SVHH106 pKa = 5.9KK107 pKa = 9.82PGKK110 pKa = 6.56TTVPRR115 pKa = 11.84IINPRR120 pKa = 11.84SPQFNILLGRR130 pKa = 11.84YY131 pKa = 7.03LTPIEE136 pKa = 4.03HH137 pKa = 6.67QIFEE141 pKa = 4.63GLQEE145 pKa = 4.25MLEE148 pKa = 4.31SPHH151 pKa = 6.38PVIAKK156 pKa = 10.17GLTQSEE162 pKa = 4.43KK163 pKa = 10.61GQIIADD169 pKa = 3.84IVNDD173 pKa = 4.82GYY175 pKa = 11.72VVVGLDD181 pKa = 3.28ASRR184 pKa = 11.84FDD186 pKa = 3.41QCISEE191 pKa = 4.54EE192 pKa = 4.05LLKK195 pKa = 10.24MEE197 pKa = 4.75HH198 pKa = 6.26NVYY201 pKa = 10.33LKK203 pKa = 10.53CYY205 pKa = 10.1QNDD208 pKa = 3.66RR209 pKa = 11.84LLRR212 pKa = 11.84ALLKK216 pKa = 10.75CQLDD220 pKa = 3.36NSGQYY225 pKa = 9.39IGRR228 pKa = 11.84DD229 pKa = 3.15GRR231 pKa = 11.84VKK233 pKa = 10.4VRR235 pKa = 11.84YY236 pKa = 8.79GAIRR240 pKa = 11.84CSGDD244 pKa = 3.03MNTSLGNCIISVLLSVLFCDD264 pKa = 4.17EE265 pKa = 4.24NGIGDD270 pKa = 3.87FRR272 pKa = 11.84VFCDD276 pKa = 4.84GDD278 pKa = 4.02DD279 pKa = 3.74LLLAVRR285 pKa = 11.84RR286 pKa = 11.84GDD288 pKa = 3.91LNKK291 pKa = 10.47LNPLQQWYY299 pKa = 8.46LQWGLRR305 pKa = 11.84MKK307 pKa = 10.59VEE309 pKa = 4.41EE310 pKa = 4.05PAYY313 pKa = 9.33TPEE316 pKa = 4.14AVEE319 pKa = 4.84FCQSRR324 pKa = 11.84PVCIDD329 pKa = 2.59GRR331 pKa = 11.84YY332 pKa = 9.67VLIRR336 pKa = 11.84DD337 pKa = 3.25VRR339 pKa = 11.84KK340 pKa = 10.17CINVDD345 pKa = 2.98YY346 pKa = 11.18SGFVNLQDD354 pKa = 3.38RR355 pKa = 11.84DD356 pKa = 3.78YY357 pKa = 9.72MLKK360 pKa = 9.51YY361 pKa = 9.9LRR363 pKa = 11.84AVGVCGSYY371 pKa = 10.34LAAGCPVLQAWYY383 pKa = 9.67GFGVRR388 pKa = 11.84VGTTGKK394 pKa = 10.53LDD396 pKa = 3.45VLDD399 pKa = 3.98TEE401 pKa = 5.26RR402 pKa = 11.84GFTRR406 pKa = 11.84QAKK409 pKa = 9.67LEE411 pKa = 4.03QRR413 pKa = 11.84SGAVMYY419 pKa = 10.49RR420 pKa = 11.84PIDD423 pKa = 4.08DD424 pKa = 3.97GARR427 pKa = 11.84EE428 pKa = 4.18SFRR431 pKa = 11.84LAFGIGAAEE440 pKa = 3.81QLLLEE445 pKa = 4.74SIFDD449 pKa = 3.6EE450 pKa = 4.38MTLEE454 pKa = 4.27RR455 pKa = 11.84GQLDD459 pKa = 3.47SHH461 pKa = 6.13PQEE464 pKa = 3.84ITDD467 pKa = 4.17FSNTFAFLVQPQHH480 pKa = 5.65GG481 pKa = 3.65
Molecular weight: 54.59 kDa
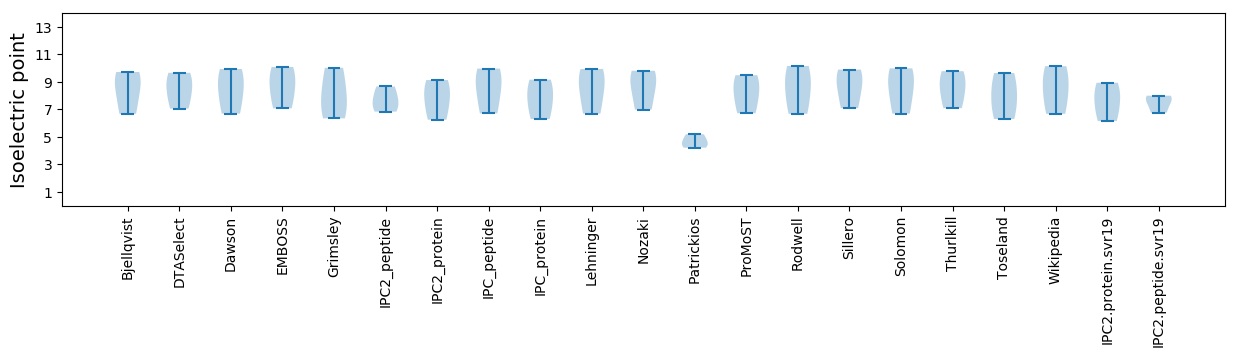
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGK5|A0A1L3KGK5_9VIRU RNA-directed RNA polymerase OS=Sanxia tombus-like virus 4 OX=1923388 PE=4 SV=1
MM1 pKa = 7.26VKK3 pKa = 10.52LSNRR7 pKa = 11.84LVGKK11 pKa = 10.48LIDD14 pKa = 3.76RR15 pKa = 11.84ASGQRR20 pKa = 11.84PRR22 pKa = 11.84NGRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84VKK29 pKa = 10.15RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84AAPRR36 pKa = 11.84GKK38 pKa = 9.54AATMRR43 pKa = 11.84RR44 pKa = 11.84EE45 pKa = 3.53IHH47 pKa = 6.13VPHH50 pKa = 6.89ANVLANPSTAPLHH63 pKa = 5.72GCVGIYY69 pKa = 10.0EE70 pKa = 4.47GEE72 pKa = 4.01QGNITRR78 pKa = 11.84LSLDD82 pKa = 3.32YY83 pKa = 10.7TIGGGAGNTCAYY95 pKa = 10.67YY96 pKa = 10.03IFHH99 pKa = 7.77PNTGLVSGAQTTAGSTAIVPAFGVGAGTSPGYY131 pKa = 10.77NLLNTNARR139 pKa = 11.84KK140 pKa = 9.88VRR142 pKa = 11.84GIAAGMRR149 pKa = 11.84FSIPSLSLTTIVGEE163 pKa = 4.32VCVGVCSADD172 pKa = 2.99TFMNVPNFTVDD183 pKa = 3.36QLFLHH188 pKa = 6.16SAGRR192 pKa = 11.84GPINKK197 pKa = 8.12ITQDD201 pKa = 3.77CAWYY205 pKa = 8.96PGSFDD210 pKa = 4.35ARR212 pKa = 11.84YY213 pKa = 8.1STYY216 pKa = 11.0AGTIADD222 pKa = 4.09TGSDD226 pKa = 3.4LSDD229 pKa = 3.14TNVFYY234 pKa = 10.15FAIRR238 pKa = 11.84GIPVSTNVQVKK249 pKa = 10.28LDD251 pKa = 3.98WVCEE255 pKa = 4.18WVAKK259 pKa = 10.57AGTGLVPSYY268 pKa = 10.39TSNPGSNHH276 pKa = 3.87QTTVAHH282 pKa = 6.18LHH284 pKa = 6.05EE285 pKa = 4.53NHH287 pKa = 7.01PGWFHH292 pKa = 5.57SVKK295 pKa = 10.62SEE297 pKa = 3.95AEE299 pKa = 3.92AVGKK303 pKa = 10.39NIVRR307 pKa = 11.84EE308 pKa = 4.19GVKK311 pKa = 10.04AARR314 pKa = 11.84HH315 pKa = 5.28KK316 pKa = 10.67AEE318 pKa = 3.75KK319 pKa = 9.14WVPTMLEE326 pKa = 3.67SGFAAFGLL334 pKa = 4.17
MM1 pKa = 7.26VKK3 pKa = 10.52LSNRR7 pKa = 11.84LVGKK11 pKa = 10.48LIDD14 pKa = 3.76RR15 pKa = 11.84ASGQRR20 pKa = 11.84PRR22 pKa = 11.84NGRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84VKK29 pKa = 10.15RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84AAPRR36 pKa = 11.84GKK38 pKa = 9.54AATMRR43 pKa = 11.84RR44 pKa = 11.84EE45 pKa = 3.53IHH47 pKa = 6.13VPHH50 pKa = 6.89ANVLANPSTAPLHH63 pKa = 5.72GCVGIYY69 pKa = 10.0EE70 pKa = 4.47GEE72 pKa = 4.01QGNITRR78 pKa = 11.84LSLDD82 pKa = 3.32YY83 pKa = 10.7TIGGGAGNTCAYY95 pKa = 10.67YY96 pKa = 10.03IFHH99 pKa = 7.77PNTGLVSGAQTTAGSTAIVPAFGVGAGTSPGYY131 pKa = 10.77NLLNTNARR139 pKa = 11.84KK140 pKa = 9.88VRR142 pKa = 11.84GIAAGMRR149 pKa = 11.84FSIPSLSLTTIVGEE163 pKa = 4.32VCVGVCSADD172 pKa = 2.99TFMNVPNFTVDD183 pKa = 3.36QLFLHH188 pKa = 6.16SAGRR192 pKa = 11.84GPINKK197 pKa = 8.12ITQDD201 pKa = 3.77CAWYY205 pKa = 8.96PGSFDD210 pKa = 4.35ARR212 pKa = 11.84YY213 pKa = 8.1STYY216 pKa = 11.0AGTIADD222 pKa = 4.09TGSDD226 pKa = 3.4LSDD229 pKa = 3.14TNVFYY234 pKa = 10.15FAIRR238 pKa = 11.84GIPVSTNVQVKK249 pKa = 10.28LDD251 pKa = 3.98WVCEE255 pKa = 4.18WVAKK259 pKa = 10.57AGTGLVPSYY268 pKa = 10.39TSNPGSNHH276 pKa = 3.87QTTVAHH282 pKa = 6.18LHH284 pKa = 6.05EE285 pKa = 4.53NHH287 pKa = 7.01PGWFHH292 pKa = 5.57SVKK295 pKa = 10.62SEE297 pKa = 3.95AEE299 pKa = 3.92AVGKK303 pKa = 10.39NIVRR307 pKa = 11.84EE308 pKa = 4.19GVKK311 pKa = 10.04AARR314 pKa = 11.84HH315 pKa = 5.28KK316 pKa = 10.67AEE318 pKa = 3.75KK319 pKa = 9.14WVPTMLEE326 pKa = 3.67SGFAAFGLL334 pKa = 4.17
Molecular weight: 35.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1026 |
211 |
481 |
342.0 |
38.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.602 ± 1.394 | 2.339 ± 0.323 |
5.556 ± 1.078 | 5.653 ± 0.982 |
4.191 ± 0.413 | 9.064 ± 0.796 |
2.242 ± 0.471 | 4.971 ± 0.266 |
5.166 ± 0.927 | 7.895 ± 1.686 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.242 ± 0.336 | 4.581 ± 0.436 |
3.996 ± 0.599 | 3.021 ± 1.046 |
7.7 ± 0.495 | 5.166 ± 0.749 |
5.556 ± 0.908 | 8.382 ± 0.711 |
1.17 ± 0.191 | 3.509 ± 0.234 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
