
Porphyra umbilicalis (Purple laver) (Red alga)
Taxonomy: cellular organisms; Eukaryota; Rhodophyta; Bangiophyceae; Bangiales; Bangiaceae; Porphyra
Average proteome isoelectric point is 7.87
Get precalculated fractions of proteins
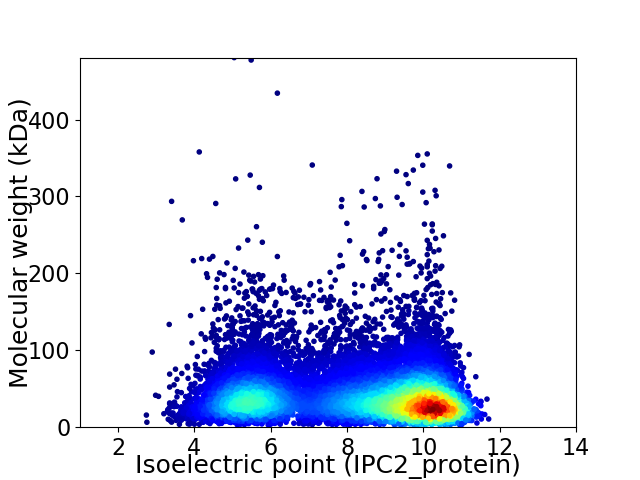
Virtual 2D-PAGE plot for 13469 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X6PII2|A0A1X6PII2_PORUM Uncharacterized protein OS=Porphyra umbilicalis OX=2786 GN=BU14_0047s0012 PE=4 SV=1
MM1 pKa = 6.73ATAVAFPLAAAVSAAAADD19 pKa = 3.75ARR21 pKa = 11.84AAPVGRR27 pKa = 11.84QAEE30 pKa = 4.74GEE32 pKa = 4.07AAPAPEE38 pKa = 4.87GGVVPLGGPCATAADD53 pKa = 4.47CAAPTDD59 pKa = 3.98GGPPAEE65 pKa = 4.58CVAGPGGDD73 pKa = 3.43GGGDD77 pKa = 3.48GDD79 pKa = 4.86GGSDD83 pKa = 3.88GDD85 pKa = 4.14GGGDD89 pKa = 3.67GDD91 pKa = 5.13SGGDD95 pKa = 3.46GDD97 pKa = 5.51GGGDD101 pKa = 3.66GDD103 pKa = 4.46GGGDD107 pKa = 3.36GGGDD111 pKa = 3.33GDD113 pKa = 4.36SDD115 pKa = 3.82NGVGDD120 pKa = 3.79EE121 pKa = 4.27VSTCRR126 pKa = 11.84RR127 pKa = 11.84RR128 pKa = 11.84HH129 pKa = 5.3AAGAPCTTAADD140 pKa = 3.68ACHH143 pKa = 6.92RR144 pKa = 11.84DD145 pKa = 3.79LLCGEE150 pKa = 5.0GGVCVPPPPEE160 pKa = 3.8EE161 pKa = 3.77AAGADD166 pKa = 3.97RR167 pKa = 11.84YY168 pKa = 10.74RR169 pKa = 11.84QFF171 pKa = 3.93
MM1 pKa = 6.73ATAVAFPLAAAVSAAAADD19 pKa = 3.75ARR21 pKa = 11.84AAPVGRR27 pKa = 11.84QAEE30 pKa = 4.74GEE32 pKa = 4.07AAPAPEE38 pKa = 4.87GGVVPLGGPCATAADD53 pKa = 4.47CAAPTDD59 pKa = 3.98GGPPAEE65 pKa = 4.58CVAGPGGDD73 pKa = 3.43GGGDD77 pKa = 3.48GDD79 pKa = 4.86GGSDD83 pKa = 3.88GDD85 pKa = 4.14GGGDD89 pKa = 3.67GDD91 pKa = 5.13SGGDD95 pKa = 3.46GDD97 pKa = 5.51GGGDD101 pKa = 3.66GDD103 pKa = 4.46GGGDD107 pKa = 3.36GGGDD111 pKa = 3.33GDD113 pKa = 4.36SDD115 pKa = 3.82NGVGDD120 pKa = 3.79EE121 pKa = 4.27VSTCRR126 pKa = 11.84RR127 pKa = 11.84RR128 pKa = 11.84HH129 pKa = 5.3AAGAPCTTAADD140 pKa = 3.68ACHH143 pKa = 6.92RR144 pKa = 11.84DD145 pKa = 3.79LLCGEE150 pKa = 5.0GGVCVPPPPEE160 pKa = 3.8EE161 pKa = 3.77AAGADD166 pKa = 3.97RR167 pKa = 11.84YY168 pKa = 10.74RR169 pKa = 11.84QFF171 pKa = 3.93
Molecular weight: 15.63 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X6NRX2|A0A1X6NRX2_PORUM Uncharacterized protein OS=Porphyra umbilicalis OX=2786 GN=BU14_0553s0002 PE=4 SV=1
MM1 pKa = 6.85TWRR4 pKa = 11.84GRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84LRR10 pKa = 11.84PPPRR14 pKa = 11.84RR15 pKa = 11.84PPSRR19 pKa = 11.84GWPNSAPRR27 pKa = 11.84ACASGAPPTTLRR39 pKa = 11.84KK40 pKa = 7.65WSKK43 pKa = 9.15RR44 pKa = 11.84TRR46 pKa = 11.84TWRR49 pKa = 11.84ASRR52 pKa = 11.84RR53 pKa = 11.84GCSTKK58 pKa = 10.08RR59 pKa = 11.84RR60 pKa = 11.84ASRR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84SSGGTTARR73 pKa = 11.84RR74 pKa = 11.84GGARR78 pKa = 11.84RR79 pKa = 11.84RR80 pKa = 11.84RR81 pKa = 11.84ARR83 pKa = 11.84RR84 pKa = 11.84RR85 pKa = 11.84WRR87 pKa = 11.84WSRR90 pKa = 11.84RR91 pKa = 11.84APRR94 pKa = 11.84APATPLRR101 pKa = 11.84PSRR104 pKa = 11.84RR105 pKa = 11.84CARR108 pKa = 11.84GGWRR112 pKa = 11.84ARR114 pKa = 11.84PRR116 pKa = 11.84RR117 pKa = 11.84RR118 pKa = 11.84RR119 pKa = 11.84RR120 pKa = 11.84RR121 pKa = 11.84RR122 pKa = 11.84RR123 pKa = 11.84RR124 pKa = 11.84WVPRR128 pKa = 11.84ARR130 pKa = 11.84RR131 pKa = 11.84KK132 pKa = 9.59RR133 pKa = 11.84ASAMAGGGRR142 pKa = 11.84AWRR145 pKa = 11.84RR146 pKa = 11.84ASALTRR152 pKa = 11.84TFPWSSSTLRR162 pKa = 11.84SSARR166 pKa = 11.84TPSLCPRR173 pKa = 11.84AARR176 pKa = 11.84GGVAAARR183 pKa = 11.84PVGRR187 pKa = 11.84RR188 pKa = 11.84GAPRR192 pKa = 11.84PATSTRR198 pKa = 11.84GRR200 pKa = 11.84TGGGGSRR207 pKa = 11.84GQAA210 pKa = 2.96
MM1 pKa = 6.85TWRR4 pKa = 11.84GRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84LRR10 pKa = 11.84PPPRR14 pKa = 11.84RR15 pKa = 11.84PPSRR19 pKa = 11.84GWPNSAPRR27 pKa = 11.84ACASGAPPTTLRR39 pKa = 11.84KK40 pKa = 7.65WSKK43 pKa = 9.15RR44 pKa = 11.84TRR46 pKa = 11.84TWRR49 pKa = 11.84ASRR52 pKa = 11.84RR53 pKa = 11.84GCSTKK58 pKa = 10.08RR59 pKa = 11.84RR60 pKa = 11.84ASRR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84SSGGTTARR73 pKa = 11.84RR74 pKa = 11.84GGARR78 pKa = 11.84RR79 pKa = 11.84RR80 pKa = 11.84RR81 pKa = 11.84ARR83 pKa = 11.84RR84 pKa = 11.84RR85 pKa = 11.84WRR87 pKa = 11.84WSRR90 pKa = 11.84RR91 pKa = 11.84APRR94 pKa = 11.84APATPLRR101 pKa = 11.84PSRR104 pKa = 11.84RR105 pKa = 11.84CARR108 pKa = 11.84GGWRR112 pKa = 11.84ARR114 pKa = 11.84PRR116 pKa = 11.84RR117 pKa = 11.84RR118 pKa = 11.84RR119 pKa = 11.84RR120 pKa = 11.84RR121 pKa = 11.84RR122 pKa = 11.84RR123 pKa = 11.84RR124 pKa = 11.84WVPRR128 pKa = 11.84ARR130 pKa = 11.84RR131 pKa = 11.84KK132 pKa = 9.59RR133 pKa = 11.84ASAMAGGGRR142 pKa = 11.84AWRR145 pKa = 11.84RR146 pKa = 11.84ASALTRR152 pKa = 11.84TFPWSSSTLRR162 pKa = 11.84SSARR166 pKa = 11.84TPSLCPRR173 pKa = 11.84AARR176 pKa = 11.84GGVAAARR183 pKa = 11.84PVGRR187 pKa = 11.84RR188 pKa = 11.84GAPRR192 pKa = 11.84PATSTRR198 pKa = 11.84GRR200 pKa = 11.84TGGGGSRR207 pKa = 11.84GQAA210 pKa = 2.96
Molecular weight: 23.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5967975 |
29 |
4583 |
443.1 |
45.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
17.515 ± 0.056 | 1.519 ± 0.011 |
4.632 ± 0.021 | 3.541 ± 0.022 |
2.029 ± 0.013 | 11.735 ± 0.05 |
2.079 ± 0.01 | 1.832 ± 0.014 |
2.119 ± 0.02 | 7.097 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.454 ± 0.009 | 1.568 ± 0.011 |
9.17 ± 0.046 | 2.058 ± 0.014 |
9.777 ± 0.049 | 6.523 ± 0.028 |
5.393 ± 0.016 | 7.315 ± 0.025 |
1.297 ± 0.009 | 1.346 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
