
Cognatishimia maritima
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Cognatishimia
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins
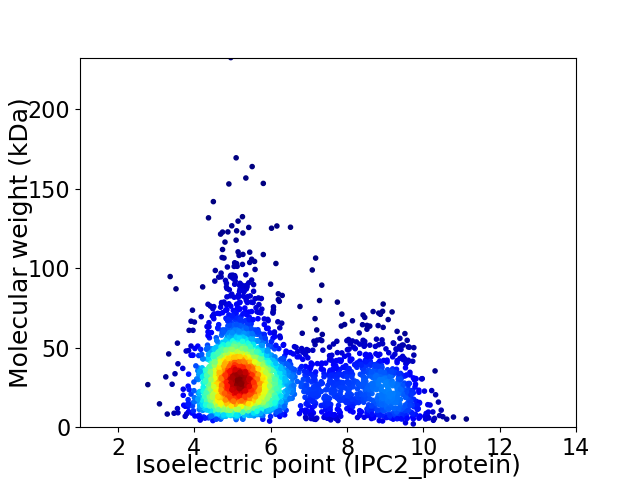
Virtual 2D-PAGE plot for 3233 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
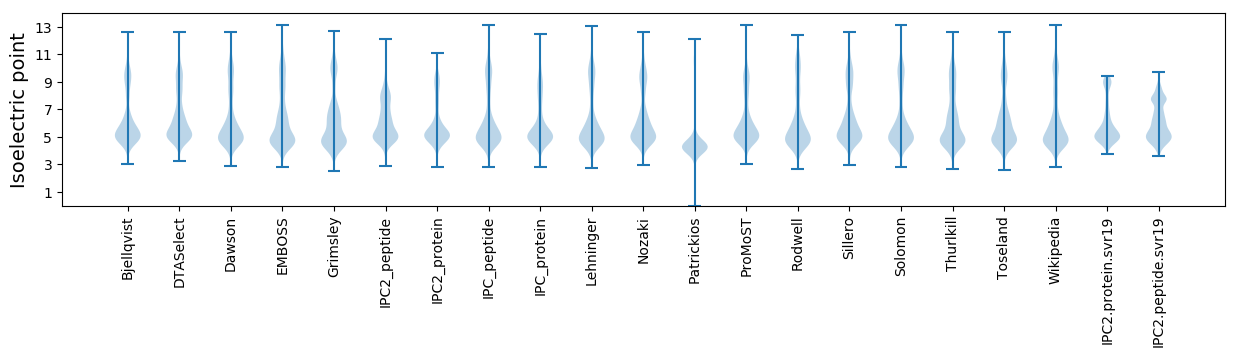
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M5TV48|A0A1M5TV48_9RHOB Acyl transferase domain-containing protein OS=Cognatishimia maritima OX=870908 GN=SAMN04488044_2709 PE=3 SV=1
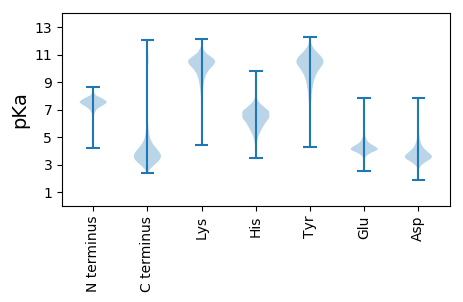
VV1 pKa = 6.71GVSGQDD7 pKa = 3.17ANGTQGVAEE16 pKa = 4.56ALNFGNFDD24 pKa = 4.22NIDD27 pKa = 3.11ITGYY31 pKa = 10.02KK32 pKa = 8.56WADD35 pKa = 3.07INGNGVWDD43 pKa = 4.05EE44 pKa = 4.4GEE46 pKa = 4.02KK47 pKa = 10.91GLNDD51 pKa = 3.2FVIYY55 pKa = 10.64LDD57 pKa = 3.72TDD59 pKa = 3.54EE60 pKa = 6.19DD61 pKa = 3.98PTNGFIRR68 pKa = 11.84MMTTEE73 pKa = 4.12NDD75 pKa = 3.16GTYY78 pKa = 10.85DD79 pKa = 3.37GFYY82 pKa = 10.31SFQDD86 pKa = 3.39VTPAEE91 pKa = 4.35INGATTLYY99 pKa = 10.15VYY101 pKa = 10.61EE102 pKa = 4.49GVEE105 pKa = 3.74PGYY108 pKa = 7.75TQTYY112 pKa = 8.03GGYY115 pKa = 9.9SIDD118 pKa = 3.78VASGAVVEE126 pKa = 4.46GTYY129 pKa = 10.35RR130 pKa = 11.84EE131 pKa = 4.27TEE133 pKa = 4.06DD134 pKa = 4.34GNFGNHH140 pKa = 5.17MMEE143 pKa = 4.79GANRR147 pKa = 11.84TPGFWQSTLGKK158 pKa = 9.55SLYY161 pKa = 10.89DD162 pKa = 3.95GDD164 pKa = 5.23PDD166 pKa = 4.05NQGDD170 pKa = 4.1ANGDD174 pKa = 3.99GIPDD178 pKa = 3.59GNKK181 pKa = 10.22DD182 pKa = 3.54FEE184 pKa = 4.48EE185 pKa = 4.62EE186 pKa = 4.25GWSEE190 pKa = 3.84EE191 pKa = 4.19DD192 pKa = 4.21LLIKK196 pKa = 10.7YY197 pKa = 10.13GIDD200 pKa = 3.45LEE202 pKa = 5.29GGDD205 pKa = 5.05GINDD209 pKa = 3.66HH210 pKa = 6.44FLVWDD215 pKa = 3.81EE216 pKa = 4.54NGNGVLDD223 pKa = 4.43GDD225 pKa = 5.09DD226 pKa = 3.12IALTFEE232 pKa = 4.61EE233 pKa = 4.45LCGWVEE239 pKa = 4.11GGGTGKK245 pKa = 9.95RR246 pKa = 11.84DD247 pKa = 3.6YY248 pKa = 11.69VEE250 pKa = 3.93VLQRR254 pKa = 11.84DD255 pKa = 3.63VGATFLNLLNNSSLAGSDD273 pKa = 3.66DD274 pKa = 4.04VIMDD278 pKa = 3.66GDD280 pKa = 3.86IEE282 pKa = 4.45DD283 pKa = 4.17SYY285 pKa = 11.37EE286 pKa = 3.8AAIMFIMDD294 pKa = 3.72NEE296 pKa = 4.35MKK298 pKa = 10.46GSKK301 pKa = 10.06KK302 pKa = 10.59AMQKK306 pKa = 9.91DD307 pKa = 3.08WKK309 pKa = 10.28DD310 pKa = 3.65YY311 pKa = 11.03GSAAHH316 pKa = 6.7NEE318 pKa = 3.28LDD320 pKa = 3.53AYY322 pKa = 10.82NNNGQVFDD330 pKa = 4.5AEE332 pKa = 4.52SGTFVQTVMDD342 pKa = 5.07GDD344 pKa = 4.36DD345 pKa = 4.22LSKK348 pKa = 9.8QTAQNYY354 pKa = 9.34FDD356 pKa = 5.76AEE358 pKa = 3.91DD359 pKa = 4.06AYY361 pKa = 10.54EE362 pKa = 4.28LFVAA366 pKa = 5.48
VV1 pKa = 6.71GVSGQDD7 pKa = 3.17ANGTQGVAEE16 pKa = 4.56ALNFGNFDD24 pKa = 4.22NIDD27 pKa = 3.11ITGYY31 pKa = 10.02KK32 pKa = 8.56WADD35 pKa = 3.07INGNGVWDD43 pKa = 4.05EE44 pKa = 4.4GEE46 pKa = 4.02KK47 pKa = 10.91GLNDD51 pKa = 3.2FVIYY55 pKa = 10.64LDD57 pKa = 3.72TDD59 pKa = 3.54EE60 pKa = 6.19DD61 pKa = 3.98PTNGFIRR68 pKa = 11.84MMTTEE73 pKa = 4.12NDD75 pKa = 3.16GTYY78 pKa = 10.85DD79 pKa = 3.37GFYY82 pKa = 10.31SFQDD86 pKa = 3.39VTPAEE91 pKa = 4.35INGATTLYY99 pKa = 10.15VYY101 pKa = 10.61EE102 pKa = 4.49GVEE105 pKa = 3.74PGYY108 pKa = 7.75TQTYY112 pKa = 8.03GGYY115 pKa = 9.9SIDD118 pKa = 3.78VASGAVVEE126 pKa = 4.46GTYY129 pKa = 10.35RR130 pKa = 11.84EE131 pKa = 4.27TEE133 pKa = 4.06DD134 pKa = 4.34GNFGNHH140 pKa = 5.17MMEE143 pKa = 4.79GANRR147 pKa = 11.84TPGFWQSTLGKK158 pKa = 9.55SLYY161 pKa = 10.89DD162 pKa = 3.95GDD164 pKa = 5.23PDD166 pKa = 4.05NQGDD170 pKa = 4.1ANGDD174 pKa = 3.99GIPDD178 pKa = 3.59GNKK181 pKa = 10.22DD182 pKa = 3.54FEE184 pKa = 4.48EE185 pKa = 4.62EE186 pKa = 4.25GWSEE190 pKa = 3.84EE191 pKa = 4.19DD192 pKa = 4.21LLIKK196 pKa = 10.7YY197 pKa = 10.13GIDD200 pKa = 3.45LEE202 pKa = 5.29GGDD205 pKa = 5.05GINDD209 pKa = 3.66HH210 pKa = 6.44FLVWDD215 pKa = 3.81EE216 pKa = 4.54NGNGVLDD223 pKa = 4.43GDD225 pKa = 5.09DD226 pKa = 3.12IALTFEE232 pKa = 4.61EE233 pKa = 4.45LCGWVEE239 pKa = 4.11GGGTGKK245 pKa = 9.95RR246 pKa = 11.84DD247 pKa = 3.6YY248 pKa = 11.69VEE250 pKa = 3.93VLQRR254 pKa = 11.84DD255 pKa = 3.63VGATFLNLLNNSSLAGSDD273 pKa = 3.66DD274 pKa = 4.04VIMDD278 pKa = 3.66GDD280 pKa = 3.86IEE282 pKa = 4.45DD283 pKa = 4.17SYY285 pKa = 11.37EE286 pKa = 3.8AAIMFIMDD294 pKa = 3.72NEE296 pKa = 4.35MKK298 pKa = 10.46GSKK301 pKa = 10.06KK302 pKa = 10.59AMQKK306 pKa = 9.91DD307 pKa = 3.08WKK309 pKa = 10.28DD310 pKa = 3.65YY311 pKa = 11.03GSAAHH316 pKa = 6.7NEE318 pKa = 3.28LDD320 pKa = 3.53AYY322 pKa = 10.82NNNGQVFDD330 pKa = 4.5AEE332 pKa = 4.52SGTFVQTVMDD342 pKa = 5.07GDD344 pKa = 4.36DD345 pKa = 4.22LSKK348 pKa = 9.8QTAQNYY354 pKa = 9.34FDD356 pKa = 5.76AEE358 pKa = 3.91DD359 pKa = 4.06AYY361 pKa = 10.54EE362 pKa = 4.28LFVAA366 pKa = 5.48
Molecular weight: 39.9 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M5MMS6|A0A1M5MMS6_9RHOB 30S ribosomal protein S13 OS=Cognatishimia maritima OX=870908 GN=rpsM PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.97ALSAA44 pKa = 4.07
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.54ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.97ALSAA44 pKa = 4.07
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
981079 |
18 |
2133 |
303.5 |
33.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.526 ± 0.048 | 0.919 ± 0.014 |
6.125 ± 0.035 | 6.123 ± 0.043 |
3.94 ± 0.031 | 8.219 ± 0.044 |
2.077 ± 0.022 | 5.487 ± 0.033 |
3.91 ± 0.036 | 9.908 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.843 ± 0.02 | 2.905 ± 0.022 |
4.753 ± 0.028 | 3.44 ± 0.026 |
6.049 ± 0.037 | 5.375 ± 0.027 |
5.441 ± 0.025 | 7.289 ± 0.032 |
1.363 ± 0.018 | 2.306 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
