
Tenacibaculum jejuense
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Tenacibaculum
Average proteome isoelectric point is 6.85
Get precalculated fractions of proteins
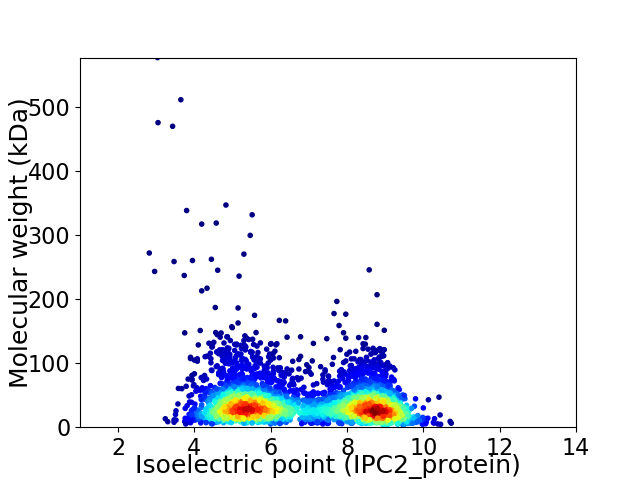
Virtual 2D-PAGE plot for 3944 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A238UEW4|A0A238UEW4_9FLAO Glycosyl transferase group 1 family protein OS=Tenacibaculum jejuense OX=584609 GN=TJEJU_4079 PE=4 SV=1
MM1 pKa = 7.88RR2 pKa = 11.84KK3 pKa = 8.09TITFLYY9 pKa = 10.15AVLISISIFGQTSLSTGDD27 pKa = 3.23IAFLSVNFDD36 pKa = 3.21GSDD39 pKa = 3.39DD40 pKa = 3.62YY41 pKa = 12.21SFILFKK47 pKa = 10.97DD48 pKa = 3.03IDD50 pKa = 4.47ANTTITFTDD59 pKa = 4.33CGWSDD64 pKa = 3.32GSASFICNVGDD75 pKa = 3.82ANGWTWLSGSALAAGTVITITNEE98 pKa = 3.35SSASIGTVSGVTSLLSSAGDD118 pKa = 3.3QIFAYY123 pKa = 9.76QGSASSPTFLAGINSNEE140 pKa = 4.21VADD143 pKa = 4.19DD144 pKa = 4.18SNWNGEE150 pKa = 4.17VTSNSTSALPDD161 pKa = 3.18QLTNGTNAIRR171 pKa = 11.84LHH173 pKa = 6.03NMGSEE178 pKa = 3.99VDD180 pKa = 3.12NWQYY184 pKa = 11.46NCTVVSGDD192 pKa = 3.37VTTVRR197 pKa = 11.84AAINNIANWSNNNGTAFSPSAPNCLWNITFASPPSTVTSVTVPSNATYY245 pKa = 10.77SAGQNLDD252 pKa = 3.34FTVSFNEE259 pKa = 4.52TITVNTTGGTPQLALTIGSTTRR281 pKa = 11.84QATFVSGSGTANLLFRR297 pKa = 11.84YY298 pKa = 6.91TVQAGDD304 pKa = 3.78SDD306 pKa = 4.03TDD308 pKa = 4.46GIAVGSLAANGGTIQNGSGTDD329 pKa = 3.33ADD331 pKa = 4.16LTLNSIGSTTNVFVDD346 pKa = 5.68GIAPTGYY353 pKa = 10.25SVTIDD358 pKa = 3.35QSPINAANDD367 pKa = 3.7DD368 pKa = 3.94AVSFTFASAEE378 pKa = 3.88VGTTYY383 pKa = 11.09NYY385 pKa = 11.26AFTSSGAPGSVTGSGTIATASDD407 pKa = 3.42QTTGIDD413 pKa = 3.27ISGLADD419 pKa = 3.41GMITLSVTLTDD430 pKa = 3.62VNGNTGAAATDD441 pKa = 3.85TEE443 pKa = 4.68TKK445 pKa = 9.33EE446 pKa = 4.32TVAPTGYY453 pKa = 10.31SVTIDD458 pKa = 3.35QSPINAANDD467 pKa = 3.7DD468 pKa = 3.94AVSFTFASAEE478 pKa = 3.88VGTTYY483 pKa = 11.09NYY485 pKa = 11.26AFTSSGAPGSVTGSGTIATASDD507 pKa = 3.98QITGIDD513 pKa = 3.26ISGLADD519 pKa = 3.31GTITLTVTLTDD530 pKa = 3.49VNGNTGAVATDD541 pKa = 3.69TEE543 pKa = 4.69TKK545 pKa = 9.32EE546 pKa = 4.32TVAPTGYY553 pKa = 10.31SVTIDD558 pKa = 3.35QSPINAANDD567 pKa = 3.66DD568 pKa = 4.04VVSFTFASAEE578 pKa = 3.88VGTTYY583 pKa = 11.09NYY585 pKa = 11.26AFTSSGAPGSVTGSGTIATASDD607 pKa = 3.98QITGIDD613 pKa = 3.26ISGLADD619 pKa = 3.37GMITLTVTLTDD630 pKa = 3.49VNGNTGAAATDD641 pKa = 3.85TEE643 pKa = 4.68TKK645 pKa = 9.33EE646 pKa = 4.32TVAPTGYY653 pKa = 10.31SVTIDD658 pKa = 3.35QSPINAANDD667 pKa = 3.7DD668 pKa = 3.94AVSFTFASAEE678 pKa = 3.88VGTTYY683 pKa = 11.09NYY685 pKa = 11.26AFTSSGAPGSVTGSGTIATASDD707 pKa = 3.98QITGIDD713 pKa = 3.26ISGLADD719 pKa = 3.31GTITLTVTLTDD730 pKa = 3.49VNGNTGAAATDD741 pKa = 3.85TEE743 pKa = 4.68TKK745 pKa = 9.33EE746 pKa = 4.32TVAPTGYY753 pKa = 10.31SVTIDD758 pKa = 3.35QSPINAANDD767 pKa = 3.7DD768 pKa = 3.94AVSFTFASAEE778 pKa = 3.88VGTTYY783 pKa = 11.09NYY785 pKa = 11.26AFTSSGAPGSVTGSGIVATATDD807 pKa = 4.36QITGIDD813 pKa = 3.55INGLADD819 pKa = 3.5GTITLTVTLTDD830 pKa = 3.49VNGNTGAAATDD841 pKa = 4.05TEE843 pKa = 4.71TKK845 pKa = 10.29DD846 pKa = 3.53ASIPLDD852 pKa = 3.29PTINVPSGAITVTTTNQTISGNYY875 pKa = 8.5AEE877 pKa = 4.77NGITIHH883 pKa = 7.35AYY885 pKa = 9.64IDD887 pKa = 3.38TDD889 pKa = 4.0NNGTADD895 pKa = 3.58NTTSLGSAIVTGNAWSFTVNLTADD919 pKa = 3.48SVNNFVVQAEE929 pKa = 4.26DD930 pKa = 3.37ASGNTSTYY938 pKa = 10.46VDD940 pKa = 4.14VPTITQTNTITWTGNTSNSWTTAGNWDD967 pKa = 4.22TNTVPTSAANVIIPSGLTNYY987 pKa = 7.21PTISSAVTVNSIEE1000 pKa = 4.23IASGASLIANTSVTANVTYY1019 pKa = 10.64NRR1021 pKa = 11.84NLPTTNWYY1029 pKa = 9.84LVSAPVSNEE1038 pKa = 3.7TQEE1041 pKa = 5.75DD1042 pKa = 4.32IINNPSHH1049 pKa = 6.64NFATGSGSNIGIGAFTNNGANPWVYY1074 pKa = 9.83ATNTTTGPLVSGAGVSMKK1092 pKa = 10.32LAAAGDD1098 pKa = 3.75VSITGNINTTNVSFPIATGTRR1119 pKa = 11.84NNFNLIGNPYY1129 pKa = 8.4TSFVNSSTFAAANTGLLSEE1148 pKa = 4.32EE1149 pKa = 4.88TVWLWDD1155 pKa = 3.29GSQYY1159 pKa = 8.9VTYY1162 pKa = 11.13NNMSPIEE1169 pKa = 4.03VAPAQGFFVEE1179 pKa = 4.86AAGSGNVTFSTANQSHH1195 pKa = 5.47QNSDD1199 pKa = 2.98TFLRR1203 pKa = 11.84QVPNPTFEE1211 pKa = 5.75LFIQNDD1217 pKa = 3.17SDD1219 pKa = 3.66KK1220 pKa = 11.2RR1221 pKa = 11.84STKK1224 pKa = 10.07VFYY1227 pKa = 11.17ANGKK1231 pKa = 7.37TKK1233 pKa = 10.61GFDD1236 pKa = 2.94NGYY1239 pKa = 9.25DD1240 pKa = 3.14SKK1242 pKa = 10.64MFAGISEE1249 pKa = 4.17NFGIFTEE1256 pKa = 4.41LLEE1259 pKa = 4.39NNQGKK1264 pKa = 9.57KK1265 pKa = 10.16LAIQTLPNSNLEE1277 pKa = 4.17TMIVPVGIIANAGEE1291 pKa = 4.76EE1292 pKa = 4.3ITFSMSSKK1300 pKa = 10.37NLPTGVEE1307 pKa = 4.1IYY1309 pKa = 11.08LEE1311 pKa = 4.3DD1312 pKa = 3.7KK1313 pKa = 11.16VNNTMINLTEE1323 pKa = 4.25RR1324 pKa = 11.84NHH1326 pKa = 5.75TLTLKK1331 pKa = 9.65TALNASGRR1339 pKa = 11.84FYY1341 pKa = 10.85IHH1343 pKa = 5.58TTAKK1347 pKa = 10.28RR1348 pKa = 11.84LSNEE1352 pKa = 4.44DD1353 pKa = 3.7IINNLNNVSIYY1364 pKa = 10.61KK1365 pKa = 9.91SASKK1369 pKa = 10.41EE1370 pKa = 3.9VTISGLQGSGDD1381 pKa = 2.97IKK1383 pKa = 11.04VFSILGKK1390 pKa = 9.92QVGSTSINSNGTSKK1404 pKa = 10.62IEE1406 pKa = 4.49LPDD1409 pKa = 4.2LPSGIYY1415 pKa = 9.59IVRR1418 pKa = 11.84LSSEE1422 pKa = 4.1FGKK1425 pKa = 8.84LTRR1428 pKa = 11.84KK1429 pKa = 9.91IILEE1433 pKa = 3.97
MM1 pKa = 7.88RR2 pKa = 11.84KK3 pKa = 8.09TITFLYY9 pKa = 10.15AVLISISIFGQTSLSTGDD27 pKa = 3.23IAFLSVNFDD36 pKa = 3.21GSDD39 pKa = 3.39DD40 pKa = 3.62YY41 pKa = 12.21SFILFKK47 pKa = 10.97DD48 pKa = 3.03IDD50 pKa = 4.47ANTTITFTDD59 pKa = 4.33CGWSDD64 pKa = 3.32GSASFICNVGDD75 pKa = 3.82ANGWTWLSGSALAAGTVITITNEE98 pKa = 3.35SSASIGTVSGVTSLLSSAGDD118 pKa = 3.3QIFAYY123 pKa = 9.76QGSASSPTFLAGINSNEE140 pKa = 4.21VADD143 pKa = 4.19DD144 pKa = 4.18SNWNGEE150 pKa = 4.17VTSNSTSALPDD161 pKa = 3.18QLTNGTNAIRR171 pKa = 11.84LHH173 pKa = 6.03NMGSEE178 pKa = 3.99VDD180 pKa = 3.12NWQYY184 pKa = 11.46NCTVVSGDD192 pKa = 3.37VTTVRR197 pKa = 11.84AAINNIANWSNNNGTAFSPSAPNCLWNITFASPPSTVTSVTVPSNATYY245 pKa = 10.77SAGQNLDD252 pKa = 3.34FTVSFNEE259 pKa = 4.52TITVNTTGGTPQLALTIGSTTRR281 pKa = 11.84QATFVSGSGTANLLFRR297 pKa = 11.84YY298 pKa = 6.91TVQAGDD304 pKa = 3.78SDD306 pKa = 4.03TDD308 pKa = 4.46GIAVGSLAANGGTIQNGSGTDD329 pKa = 3.33ADD331 pKa = 4.16LTLNSIGSTTNVFVDD346 pKa = 5.68GIAPTGYY353 pKa = 10.25SVTIDD358 pKa = 3.35QSPINAANDD367 pKa = 3.7DD368 pKa = 3.94AVSFTFASAEE378 pKa = 3.88VGTTYY383 pKa = 11.09NYY385 pKa = 11.26AFTSSGAPGSVTGSGTIATASDD407 pKa = 3.42QTTGIDD413 pKa = 3.27ISGLADD419 pKa = 3.41GMITLSVTLTDD430 pKa = 3.62VNGNTGAAATDD441 pKa = 3.85TEE443 pKa = 4.68TKK445 pKa = 9.33EE446 pKa = 4.32TVAPTGYY453 pKa = 10.31SVTIDD458 pKa = 3.35QSPINAANDD467 pKa = 3.7DD468 pKa = 3.94AVSFTFASAEE478 pKa = 3.88VGTTYY483 pKa = 11.09NYY485 pKa = 11.26AFTSSGAPGSVTGSGTIATASDD507 pKa = 3.98QITGIDD513 pKa = 3.26ISGLADD519 pKa = 3.31GTITLTVTLTDD530 pKa = 3.49VNGNTGAVATDD541 pKa = 3.69TEE543 pKa = 4.69TKK545 pKa = 9.32EE546 pKa = 4.32TVAPTGYY553 pKa = 10.31SVTIDD558 pKa = 3.35QSPINAANDD567 pKa = 3.66DD568 pKa = 4.04VVSFTFASAEE578 pKa = 3.88VGTTYY583 pKa = 11.09NYY585 pKa = 11.26AFTSSGAPGSVTGSGTIATASDD607 pKa = 3.98QITGIDD613 pKa = 3.26ISGLADD619 pKa = 3.37GMITLTVTLTDD630 pKa = 3.49VNGNTGAAATDD641 pKa = 3.85TEE643 pKa = 4.68TKK645 pKa = 9.33EE646 pKa = 4.32TVAPTGYY653 pKa = 10.31SVTIDD658 pKa = 3.35QSPINAANDD667 pKa = 3.7DD668 pKa = 3.94AVSFTFASAEE678 pKa = 3.88VGTTYY683 pKa = 11.09NYY685 pKa = 11.26AFTSSGAPGSVTGSGTIATASDD707 pKa = 3.98QITGIDD713 pKa = 3.26ISGLADD719 pKa = 3.31GTITLTVTLTDD730 pKa = 3.49VNGNTGAAATDD741 pKa = 3.85TEE743 pKa = 4.68TKK745 pKa = 9.33EE746 pKa = 4.32TVAPTGYY753 pKa = 10.31SVTIDD758 pKa = 3.35QSPINAANDD767 pKa = 3.7DD768 pKa = 3.94AVSFTFASAEE778 pKa = 3.88VGTTYY783 pKa = 11.09NYY785 pKa = 11.26AFTSSGAPGSVTGSGIVATATDD807 pKa = 4.36QITGIDD813 pKa = 3.55INGLADD819 pKa = 3.5GTITLTVTLTDD830 pKa = 3.49VNGNTGAAATDD841 pKa = 4.05TEE843 pKa = 4.71TKK845 pKa = 10.29DD846 pKa = 3.53ASIPLDD852 pKa = 3.29PTINVPSGAITVTTTNQTISGNYY875 pKa = 8.5AEE877 pKa = 4.77NGITIHH883 pKa = 7.35AYY885 pKa = 9.64IDD887 pKa = 3.38TDD889 pKa = 4.0NNGTADD895 pKa = 3.58NTTSLGSAIVTGNAWSFTVNLTADD919 pKa = 3.48SVNNFVVQAEE929 pKa = 4.26DD930 pKa = 3.37ASGNTSTYY938 pKa = 10.46VDD940 pKa = 4.14VPTITQTNTITWTGNTSNSWTTAGNWDD967 pKa = 4.22TNTVPTSAANVIIPSGLTNYY987 pKa = 7.21PTISSAVTVNSIEE1000 pKa = 4.23IASGASLIANTSVTANVTYY1019 pKa = 10.64NRR1021 pKa = 11.84NLPTTNWYY1029 pKa = 9.84LVSAPVSNEE1038 pKa = 3.7TQEE1041 pKa = 5.75DD1042 pKa = 4.32IINNPSHH1049 pKa = 6.64NFATGSGSNIGIGAFTNNGANPWVYY1074 pKa = 9.83ATNTTTGPLVSGAGVSMKK1092 pKa = 10.32LAAAGDD1098 pKa = 3.75VSITGNINTTNVSFPIATGTRR1119 pKa = 11.84NNFNLIGNPYY1129 pKa = 8.4TSFVNSSTFAAANTGLLSEE1148 pKa = 4.32EE1149 pKa = 4.88TVWLWDD1155 pKa = 3.29GSQYY1159 pKa = 8.9VTYY1162 pKa = 11.13NNMSPIEE1169 pKa = 4.03VAPAQGFFVEE1179 pKa = 4.86AAGSGNVTFSTANQSHH1195 pKa = 5.47QNSDD1199 pKa = 2.98TFLRR1203 pKa = 11.84QVPNPTFEE1211 pKa = 5.75LFIQNDD1217 pKa = 3.17SDD1219 pKa = 3.66KK1220 pKa = 11.2RR1221 pKa = 11.84STKK1224 pKa = 10.07VFYY1227 pKa = 11.17ANGKK1231 pKa = 7.37TKK1233 pKa = 10.61GFDD1236 pKa = 2.94NGYY1239 pKa = 9.25DD1240 pKa = 3.14SKK1242 pKa = 10.64MFAGISEE1249 pKa = 4.17NFGIFTEE1256 pKa = 4.41LLEE1259 pKa = 4.39NNQGKK1264 pKa = 9.57KK1265 pKa = 10.16LAIQTLPNSNLEE1277 pKa = 4.17TMIVPVGIIANAGEE1291 pKa = 4.76EE1292 pKa = 4.3ITFSMSSKK1300 pKa = 10.37NLPTGVEE1307 pKa = 4.1IYY1309 pKa = 11.08LEE1311 pKa = 4.3DD1312 pKa = 3.7KK1313 pKa = 11.16VNNTMINLTEE1323 pKa = 4.25RR1324 pKa = 11.84NHH1326 pKa = 5.75TLTLKK1331 pKa = 9.65TALNASGRR1339 pKa = 11.84FYY1341 pKa = 10.85IHH1343 pKa = 5.58TTAKK1347 pKa = 10.28RR1348 pKa = 11.84LSNEE1352 pKa = 4.44DD1353 pKa = 3.7IINNLNNVSIYY1364 pKa = 10.61KK1365 pKa = 9.91SASKK1369 pKa = 10.41EE1370 pKa = 3.9VTISGLQGSGDD1381 pKa = 2.97IKK1383 pKa = 11.04VFSILGKK1390 pKa = 9.92QVGSTSINSNGTSKK1404 pKa = 10.62IEE1406 pKa = 4.49LPDD1409 pKa = 4.2LPSGIYY1415 pKa = 9.59IVRR1418 pKa = 11.84LSSEE1422 pKa = 4.1FGKK1425 pKa = 8.84LTRR1428 pKa = 11.84KK1429 pKa = 9.91IILEE1433 pKa = 3.97
Molecular weight: 147.25 kDa
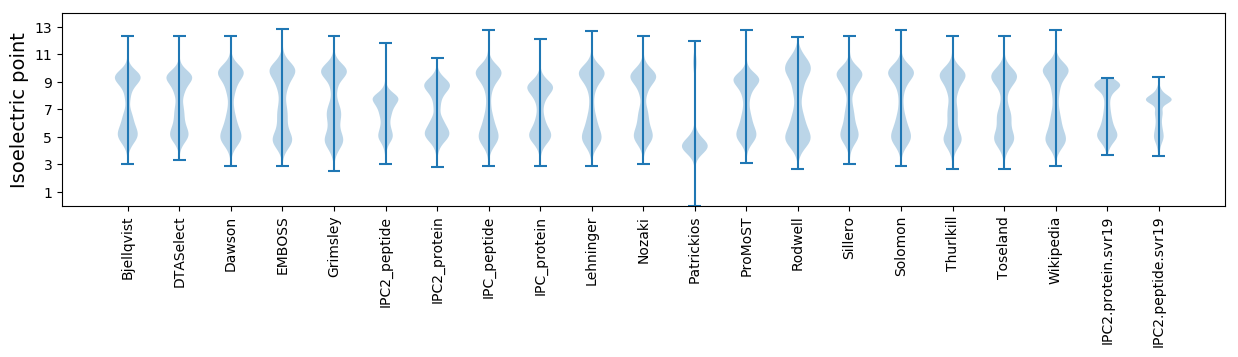
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A238U9H0|A0A238U9H0_9FLAO Probable ABC-type transport system permease component OS=Tenacibaculum jejuense OX=584609 GN=TJEJU_2160 PE=4 SV=1
MM1 pKa = 7.46KK2 pKa = 10.55FEE4 pKa = 5.36LYY6 pKa = 10.21LIKK9 pKa = 10.16IIMQSYY15 pKa = 7.54NTSLSLINTIIKK27 pKa = 8.33TDD29 pKa = 3.08NRR31 pKa = 11.84ISVRR35 pKa = 11.84RR36 pKa = 11.84FFPRR40 pKa = 11.84PP41 pKa = 3.17
MM1 pKa = 7.46KK2 pKa = 10.55FEE4 pKa = 5.36LYY6 pKa = 10.21LIKK9 pKa = 10.16IIMQSYY15 pKa = 7.54NTSLSLINTIIKK27 pKa = 8.33TDD29 pKa = 3.08NRR31 pKa = 11.84ISVRR35 pKa = 11.84RR36 pKa = 11.84FFPRR40 pKa = 11.84PP41 pKa = 3.17
Molecular weight: 4.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1390372 |
23 |
5673 |
352.5 |
39.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.574 ± 0.039 | 0.742 ± 0.015 |
5.566 ± 0.038 | 6.654 ± 0.048 |
5.396 ± 0.04 | 6.074 ± 0.055 |
1.663 ± 0.022 | 8.209 ± 0.042 |
8.528 ± 0.087 | 9.117 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.808 ± 0.018 | 6.892 ± 0.044 |
3.148 ± 0.031 | 3.23 ± 0.022 |
3.285 ± 0.03 | 6.797 ± 0.048 |
6.084 ± 0.071 | 6.122 ± 0.044 |
1.013 ± 0.014 | 4.097 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
