
Circovirus-like genome DCCV-6
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 8.59
Get precalculated fractions of proteins
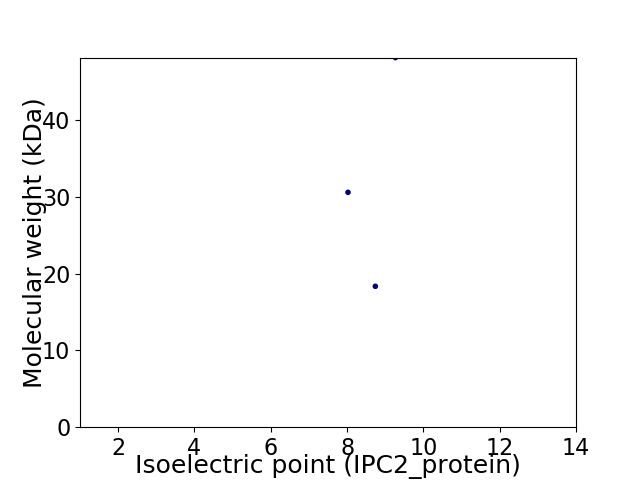
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A190WHB0|A0A190WHB0_9CIRC Uncharacterized protein OS=Circovirus-like genome DCCV-6 OX=1788446 PE=4 SV=1
MM1 pKa = 7.36TGCPPQDD8 pKa = 3.18VKK10 pKa = 11.37ALSGSGRR17 pKa = 11.84SQLISGDD24 pKa = 3.58PLYY27 pKa = 10.46TPKK30 pKa = 10.82SPTLRR35 pKa = 11.84DD36 pKa = 3.31RR37 pKa = 11.84RR38 pKa = 11.84KK39 pKa = 10.5VGLTLDD45 pKa = 3.56TCIGRR50 pKa = 11.84SSSTSKK56 pKa = 10.8RR57 pKa = 11.84KK58 pKa = 10.19DD59 pKa = 3.26PLPLSRR65 pKa = 11.84HH66 pKa = 5.62AFLQRR71 pKa = 11.84VTMNSPDD78 pKa = 3.78QLPLKK83 pKa = 10.75SMSLKK88 pKa = 9.63RR89 pKa = 11.84TLEE92 pKa = 4.03SQTLSSSSEE101 pKa = 4.21SNLSCEE107 pKa = 4.18AEE109 pKa = 4.09KK110 pKa = 10.85QIGMKK115 pKa = 10.08SGMLQEE121 pKa = 5.87AEE123 pKa = 4.41TSWLSLPMYY132 pKa = 9.88EE133 pKa = 4.61VLNFNQSNVILTSEE147 pKa = 4.35EE148 pKa = 3.82FAQILPNLSQWNVRR162 pKa = 11.84FTYY165 pKa = 10.32FGEE168 pKa = 4.18IPVLEE173 pKa = 4.45NPAGLGMRR181 pKa = 11.84RR182 pKa = 11.84VSALTLKK189 pKa = 10.89VPVQNSGMVTKK200 pKa = 10.29IKK202 pKa = 10.77KK203 pKa = 8.7MLLSMSLEE211 pKa = 4.09EE212 pKa = 4.19TSVILINKK220 pKa = 9.53ASLTCSDD227 pKa = 3.81GWTDD231 pKa = 2.92IPFWSRR237 pKa = 11.84QKK239 pKa = 11.08DD240 pKa = 3.34QGSYY244 pKa = 10.92CLLAEE249 pKa = 4.41SGSRR253 pKa = 11.84VTSRR257 pKa = 11.84LPTGILLWMRR267 pKa = 11.84TLPLLCCEE275 pKa = 4.65DD276 pKa = 3.51
MM1 pKa = 7.36TGCPPQDD8 pKa = 3.18VKK10 pKa = 11.37ALSGSGRR17 pKa = 11.84SQLISGDD24 pKa = 3.58PLYY27 pKa = 10.46TPKK30 pKa = 10.82SPTLRR35 pKa = 11.84DD36 pKa = 3.31RR37 pKa = 11.84RR38 pKa = 11.84KK39 pKa = 10.5VGLTLDD45 pKa = 3.56TCIGRR50 pKa = 11.84SSSTSKK56 pKa = 10.8RR57 pKa = 11.84KK58 pKa = 10.19DD59 pKa = 3.26PLPLSRR65 pKa = 11.84HH66 pKa = 5.62AFLQRR71 pKa = 11.84VTMNSPDD78 pKa = 3.78QLPLKK83 pKa = 10.75SMSLKK88 pKa = 9.63RR89 pKa = 11.84TLEE92 pKa = 4.03SQTLSSSSEE101 pKa = 4.21SNLSCEE107 pKa = 4.18AEE109 pKa = 4.09KK110 pKa = 10.85QIGMKK115 pKa = 10.08SGMLQEE121 pKa = 5.87AEE123 pKa = 4.41TSWLSLPMYY132 pKa = 9.88EE133 pKa = 4.61VLNFNQSNVILTSEE147 pKa = 4.35EE148 pKa = 3.82FAQILPNLSQWNVRR162 pKa = 11.84FTYY165 pKa = 10.32FGEE168 pKa = 4.18IPVLEE173 pKa = 4.45NPAGLGMRR181 pKa = 11.84RR182 pKa = 11.84VSALTLKK189 pKa = 10.89VPVQNSGMVTKK200 pKa = 10.29IKK202 pKa = 10.77KK203 pKa = 8.7MLLSMSLEE211 pKa = 4.09EE212 pKa = 4.19TSVILINKK220 pKa = 9.53ASLTCSDD227 pKa = 3.81GWTDD231 pKa = 2.92IPFWSRR237 pKa = 11.84QKK239 pKa = 11.08DD240 pKa = 3.34QGSYY244 pKa = 10.92CLLAEE249 pKa = 4.41SGSRR253 pKa = 11.84VTSRR257 pKa = 11.84LPTGILLWMRR267 pKa = 11.84TLPLLCCEE275 pKa = 4.65DD276 pKa = 3.51
Molecular weight: 30.58 kDa
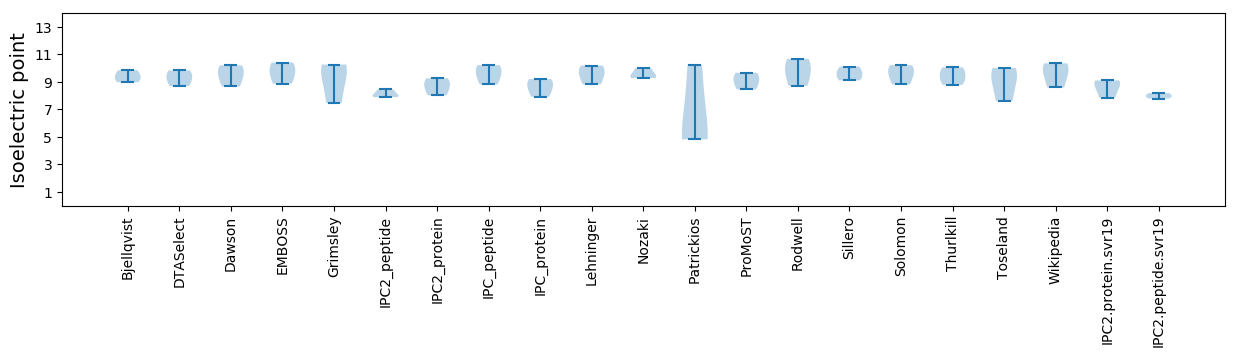
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A190WHC4|A0A190WHC4_9CIRC Viral_Rep domain-containing protein OS=Circovirus-like genome DCCV-6 OX=1788446 PE=4 SV=1
MM1 pKa = 7.24SVSRR5 pKa = 11.84TPRR8 pKa = 11.84SYY10 pKa = 11.28GGQTLVGSRR19 pKa = 11.84SRR21 pKa = 11.84GLFRR25 pKa = 11.84NTHH28 pKa = 5.38NLAVGYY34 pKa = 9.5GSRR37 pKa = 11.84LLSAGIKK44 pKa = 10.14RR45 pKa = 11.84GAQHH49 pKa = 7.29LMTKK53 pKa = 8.21ITKK56 pKa = 9.36KK57 pKa = 10.53AKK59 pKa = 9.73KK60 pKa = 9.91ALAKK64 pKa = 10.53KK65 pKa = 7.52PTGYY69 pKa = 10.85NEE71 pKa = 3.65VSRR74 pKa = 11.84SDD76 pKa = 3.27RR77 pKa = 11.84AAKK80 pKa = 10.02VINRR84 pKa = 11.84YY85 pKa = 7.78NKK87 pKa = 9.35KK88 pKa = 10.34VSVKK92 pKa = 9.74KK93 pKa = 8.45PQRR96 pKa = 11.84VKK98 pKa = 10.65VSRR101 pKa = 11.84KK102 pKa = 9.05FKK104 pKa = 10.87AKK106 pKa = 10.03VNKK109 pKa = 10.08ALEE112 pKa = 4.24PQMSKK117 pKa = 10.6GKK119 pKa = 10.47YY120 pKa = 8.54EE121 pKa = 3.92EE122 pKa = 4.31TSVFRR127 pKa = 11.84QNFGIAFGNKK137 pKa = 8.89QISGRR142 pKa = 11.84LAPDD146 pKa = 3.5TGAGSRR152 pKa = 11.84HH153 pKa = 5.91ALFGPAEE160 pKa = 4.18ILDD163 pKa = 3.95KK164 pKa = 11.18ASCLFNNRR172 pKa = 11.84VAGQTVATQTGNDD185 pKa = 3.73TNFGNASLDD194 pKa = 3.32PRR196 pKa = 11.84TAKK199 pKa = 9.03ITVNYY204 pKa = 10.14QKK206 pKa = 10.58YY207 pKa = 8.92QCTLRR212 pKa = 11.84NNSQRR217 pKa = 11.84TWIIQIYY224 pKa = 7.85EE225 pKa = 4.05ASPKK229 pKa = 10.9RR230 pKa = 11.84MMNVCADD237 pKa = 4.04ADD239 pKa = 3.91VLSQWEE245 pKa = 4.08SAMINEE251 pKa = 4.73FNNDD255 pKa = 3.13AGNAEE260 pKa = 4.23LKK262 pKa = 10.55KK263 pKa = 10.67INLANATPNTLYY275 pKa = 9.38ATPFMSKK282 pKa = 10.52AFMKK286 pKa = 10.24DD287 pKa = 2.86WNIEE291 pKa = 3.8KK292 pKa = 10.13HH293 pKa = 7.13DD294 pKa = 4.86IILDD298 pKa = 3.73PGQVYY303 pKa = 10.37DD304 pKa = 3.86HH305 pKa = 6.58VMYY308 pKa = 10.77GPTGTYY314 pKa = 10.44DD315 pKa = 3.66FSKK318 pKa = 10.04FWRR321 pKa = 11.84SDD323 pKa = 2.95GVGASNEE330 pKa = 4.2FYY332 pKa = 11.03NLQPKK337 pKa = 7.5FTRR340 pKa = 11.84EE341 pKa = 3.63LFYY344 pKa = 10.01TARR347 pKa = 11.84LDD349 pKa = 4.21LCATDD354 pKa = 3.65TGAVGRR360 pKa = 11.84WGINDD365 pKa = 3.53TTTGLIHH372 pKa = 7.12EE373 pKa = 4.46MTKK376 pKa = 8.92TCSISVPQQAGGVLTEE392 pKa = 4.38TGGLTAFRR400 pKa = 11.84NTQQRR405 pKa = 11.84DD406 pKa = 3.57AYY408 pKa = 9.98IKK410 pKa = 10.18IHH412 pKa = 5.5WNEE415 pKa = 3.64NATAVPTVRR424 pKa = 11.84VDD426 pKa = 3.24VQTAGDD432 pKa = 3.72NLL434 pKa = 3.87
MM1 pKa = 7.24SVSRR5 pKa = 11.84TPRR8 pKa = 11.84SYY10 pKa = 11.28GGQTLVGSRR19 pKa = 11.84SRR21 pKa = 11.84GLFRR25 pKa = 11.84NTHH28 pKa = 5.38NLAVGYY34 pKa = 9.5GSRR37 pKa = 11.84LLSAGIKK44 pKa = 10.14RR45 pKa = 11.84GAQHH49 pKa = 7.29LMTKK53 pKa = 8.21ITKK56 pKa = 9.36KK57 pKa = 10.53AKK59 pKa = 9.73KK60 pKa = 9.91ALAKK64 pKa = 10.53KK65 pKa = 7.52PTGYY69 pKa = 10.85NEE71 pKa = 3.65VSRR74 pKa = 11.84SDD76 pKa = 3.27RR77 pKa = 11.84AAKK80 pKa = 10.02VINRR84 pKa = 11.84YY85 pKa = 7.78NKK87 pKa = 9.35KK88 pKa = 10.34VSVKK92 pKa = 9.74KK93 pKa = 8.45PQRR96 pKa = 11.84VKK98 pKa = 10.65VSRR101 pKa = 11.84KK102 pKa = 9.05FKK104 pKa = 10.87AKK106 pKa = 10.03VNKK109 pKa = 10.08ALEE112 pKa = 4.24PQMSKK117 pKa = 10.6GKK119 pKa = 10.47YY120 pKa = 8.54EE121 pKa = 3.92EE122 pKa = 4.31TSVFRR127 pKa = 11.84QNFGIAFGNKK137 pKa = 8.89QISGRR142 pKa = 11.84LAPDD146 pKa = 3.5TGAGSRR152 pKa = 11.84HH153 pKa = 5.91ALFGPAEE160 pKa = 4.18ILDD163 pKa = 3.95KK164 pKa = 11.18ASCLFNNRR172 pKa = 11.84VAGQTVATQTGNDD185 pKa = 3.73TNFGNASLDD194 pKa = 3.32PRR196 pKa = 11.84TAKK199 pKa = 9.03ITVNYY204 pKa = 10.14QKK206 pKa = 10.58YY207 pKa = 8.92QCTLRR212 pKa = 11.84NNSQRR217 pKa = 11.84TWIIQIYY224 pKa = 7.85EE225 pKa = 4.05ASPKK229 pKa = 10.9RR230 pKa = 11.84MMNVCADD237 pKa = 4.04ADD239 pKa = 3.91VLSQWEE245 pKa = 4.08SAMINEE251 pKa = 4.73FNNDD255 pKa = 3.13AGNAEE260 pKa = 4.23LKK262 pKa = 10.55KK263 pKa = 10.67INLANATPNTLYY275 pKa = 9.38ATPFMSKK282 pKa = 10.52AFMKK286 pKa = 10.24DD287 pKa = 2.86WNIEE291 pKa = 3.8KK292 pKa = 10.13HH293 pKa = 7.13DD294 pKa = 4.86IILDD298 pKa = 3.73PGQVYY303 pKa = 10.37DD304 pKa = 3.86HH305 pKa = 6.58VMYY308 pKa = 10.77GPTGTYY314 pKa = 10.44DD315 pKa = 3.66FSKK318 pKa = 10.04FWRR321 pKa = 11.84SDD323 pKa = 2.95GVGASNEE330 pKa = 4.2FYY332 pKa = 11.03NLQPKK337 pKa = 7.5FTRR340 pKa = 11.84EE341 pKa = 3.63LFYY344 pKa = 10.01TARR347 pKa = 11.84LDD349 pKa = 4.21LCATDD354 pKa = 3.65TGAVGRR360 pKa = 11.84WGINDD365 pKa = 3.53TTTGLIHH372 pKa = 7.12EE373 pKa = 4.46MTKK376 pKa = 8.92TCSISVPQQAGGVLTEE392 pKa = 4.38TGGLTAFRR400 pKa = 11.84NTQQRR405 pKa = 11.84DD406 pKa = 3.57AYY408 pKa = 9.98IKK410 pKa = 10.18IHH412 pKa = 5.5WNEE415 pKa = 3.64NATAVPTVRR424 pKa = 11.84VDD426 pKa = 3.24VQTAGDD432 pKa = 3.72NLL434 pKa = 3.87
Molecular weight: 48.13 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
870 |
160 |
434 |
290.0 |
32.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.356 ± 1.677 | 1.609 ± 0.388 |
4.368 ± 0.292 | 4.483 ± 0.656 |
3.793 ± 0.681 | 7.126 ± 0.57 |
1.264 ± 0.359 | 4.483 ± 0.195 |
7.011 ± 0.672 | 8.391 ± 2.279 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.874 ± 0.471 | 5.287 ± 1.324 |
4.598 ± 0.788 | 4.368 ± 0.278 |
6.207 ± 0.177 | 8.391 ± 2.153 |
7.586 ± 0.618 | 5.862 ± 0.475 |
1.954 ± 0.443 | 2.989 ± 0.603 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
