
Sanxia picorna-like virus 11
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
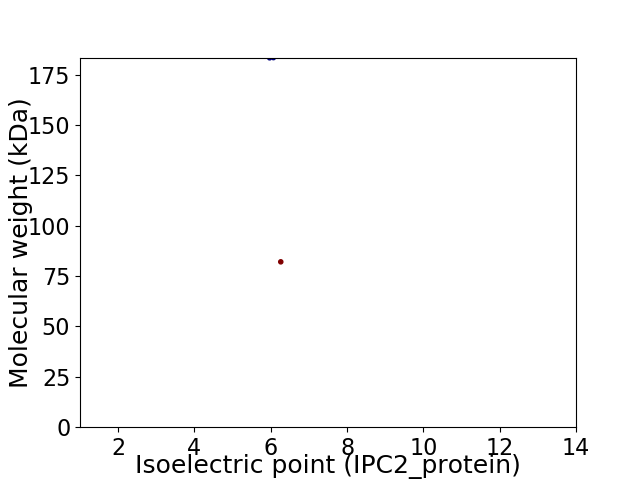
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KJC4|A0A1L3KJC4_9VIRU Uncharacterized protein OS=Sanxia picorna-like virus 11 OX=1923368 PE=4 SV=1
MM1 pKa = 8.15DD2 pKa = 3.6NQINRR7 pKa = 11.84FAPPQKK13 pKa = 8.9VHH15 pKa = 6.55AVIKK19 pKa = 6.88TTVVDD24 pKa = 4.55FRR26 pKa = 11.84VQRR29 pKa = 11.84LSRR32 pKa = 11.84KK33 pKa = 9.32YY34 pKa = 10.96LDD36 pKa = 4.22GSLTNQGAKK45 pKa = 8.78QKK47 pKa = 10.54RR48 pKa = 11.84QFDD51 pKa = 4.12RR52 pKa = 11.84LSSSDD57 pKa = 3.06KK58 pKa = 10.91RR59 pKa = 11.84EE60 pKa = 3.68WRR62 pKa = 11.84EE63 pKa = 3.43ALQRR67 pKa = 11.84AIANVKK73 pKa = 8.09NHH75 pKa = 6.75CAMQALRR82 pKa = 11.84GGKK85 pKa = 9.35LRR87 pKa = 11.84GKK89 pKa = 9.06SAHH92 pKa = 6.08RR93 pKa = 11.84FEE95 pKa = 6.15RR96 pKa = 11.84MTKK99 pKa = 10.49LEE101 pKa = 4.28AALISQFDD109 pKa = 3.69EE110 pKa = 4.01RR111 pKa = 11.84VEE113 pKa = 4.24FQSGIAVAAGSVAAASIAFASYY135 pKa = 7.36QTKK138 pKa = 10.08KK139 pKa = 10.81VSDD142 pKa = 4.01SVATAVDD149 pKa = 3.37TGNDD153 pKa = 3.61LMRR156 pKa = 11.84LLIDD160 pKa = 3.57QINGFVSKK168 pKa = 9.33ITDD171 pKa = 3.33VGSVMSKK178 pKa = 9.75IALAILLVWMMVNYY192 pKa = 9.81GHH194 pKa = 7.41LPIFVAVVLACAGSYY209 pKa = 10.25LPEE212 pKa = 6.15AIEE215 pKa = 4.4MIKK218 pKa = 10.57SIAPKK223 pKa = 10.45GISLQNGGVSLAAHH237 pKa = 7.18LLSLGCTMWIPGKK250 pKa = 9.37DD251 pKa = 3.49VKK253 pKa = 11.08SVTGEE258 pKa = 3.78FLKK261 pKa = 10.66RR262 pKa = 11.84ASYY265 pKa = 9.99FPRR268 pKa = 11.84ATEE271 pKa = 4.44GIEE274 pKa = 4.25AFMKK278 pKa = 10.43KK279 pKa = 9.75GLEE282 pKa = 3.93MFEE285 pKa = 4.14GFINFVLRR293 pKa = 11.84RR294 pKa = 11.84NEE296 pKa = 3.9EE297 pKa = 3.81SWISLQGKK305 pKa = 7.42TDD307 pKa = 3.42VFVSWKK313 pKa = 9.76RR314 pKa = 11.84KK315 pKa = 8.46VIAMLKK321 pKa = 10.0FMQEE325 pKa = 3.87QPVVPIEE332 pKa = 4.5KK333 pKa = 9.58IRR335 pKa = 11.84EE336 pKa = 4.24CKK338 pKa = 10.29DD339 pKa = 3.41LLLTGYY345 pKa = 10.55GLYY348 pKa = 9.84QVLVTQEE355 pKa = 4.3SKK357 pKa = 11.06RR358 pKa = 11.84EE359 pKa = 3.78LNFWIEE365 pKa = 3.84KK366 pKa = 9.94LAVKK370 pKa = 7.65MQPHH374 pKa = 5.6QGALQAEE381 pKa = 4.89SNIRR385 pKa = 11.84PMPYY389 pKa = 9.34FIMIGGSTGVGKK401 pKa = 9.16TSLMRR406 pKa = 11.84LIGSTILMLSGEE418 pKa = 4.56VKK420 pKa = 10.07PSEE423 pKa = 4.13ALEE426 pKa = 4.2HH427 pKa = 6.47LWQKK431 pKa = 9.09GTTEE435 pKa = 3.87YY436 pKa = 10.38WNGYY440 pKa = 8.27VGQKK444 pKa = 10.04CLIMDD449 pKa = 4.68DD450 pKa = 3.53AFQVKK455 pKa = 9.56GKK457 pKa = 9.33PGDD460 pKa = 3.66MDD462 pKa = 4.04SEE464 pKa = 4.26AMQVIRR470 pKa = 11.84AVGNWSYY477 pKa = 11.29PLNFADD483 pKa = 5.86LEE485 pKa = 4.83SKK487 pKa = 10.84GKK489 pKa = 10.44FYY491 pKa = 11.26LDD493 pKa = 3.37TPLVIGTTNSRR504 pKa = 11.84NVKK507 pKa = 10.45AEE509 pKa = 3.58WAPFITEE516 pKa = 3.8PEE518 pKa = 3.83ALVRR522 pKa = 11.84RR523 pKa = 11.84FQGSYY528 pKa = 9.46WIEE531 pKa = 3.88LNPDD535 pKa = 3.17YY536 pKa = 10.07KK537 pKa = 10.93TEE539 pKa = 4.45DD540 pKa = 3.14GRR542 pKa = 11.84FDD544 pKa = 3.4FTKK547 pKa = 10.86VNDD550 pKa = 3.8EE551 pKa = 4.16FRR553 pKa = 11.84GALQRR558 pKa = 11.84IAAMKK563 pKa = 10.37SEE565 pKa = 3.72GRR567 pKa = 11.84RR568 pKa = 11.84LSVSDD573 pKa = 5.36VMDD576 pKa = 3.46QMPWQVWTLKK586 pKa = 9.78QHH588 pKa = 6.05GFDD591 pKa = 4.1RR592 pKa = 11.84EE593 pKa = 4.3NISEE597 pKa = 4.07DD598 pKa = 3.53TAPGGLRR605 pKa = 11.84NAVEE609 pKa = 3.63IAARR613 pKa = 11.84EE614 pKa = 3.7IRR616 pKa = 11.84QRR618 pKa = 11.84RR619 pKa = 11.84EE620 pKa = 3.88SNVEE624 pKa = 3.7QIQDD628 pKa = 3.76LKK630 pKa = 11.18NWTSILGEE638 pKa = 4.29ALDD641 pKa = 5.49ADD643 pKa = 4.85LEE645 pKa = 4.51LQSGLPPSMSAFVNAMSEE663 pKa = 4.2TGISDD668 pKa = 3.59TVNRR672 pKa = 11.84RR673 pKa = 11.84LFEE676 pKa = 3.87VSTRR680 pKa = 11.84QDD682 pKa = 3.46EE683 pKa = 4.43IQEE686 pKa = 4.05EE687 pKa = 4.62SQWDD691 pKa = 3.31IPMIGVQPDD700 pKa = 4.13DD701 pKa = 3.66FVGWLSSDD709 pKa = 3.68DD710 pKa = 4.44DD711 pKa = 4.7RR712 pKa = 11.84SEE714 pKa = 4.13TQSMVDD720 pKa = 3.13GVYY723 pKa = 10.42IMPEE727 pKa = 3.83AEE729 pKa = 4.27VEE731 pKa = 3.94HH732 pKa = 7.42HH733 pKa = 6.89GILRR737 pKa = 11.84KK738 pKa = 10.05LYY740 pKa = 10.44DD741 pKa = 3.75VLTSWFDD748 pKa = 3.36KK749 pKa = 11.1LPIRR753 pKa = 11.84KK754 pKa = 8.14MVAGVLAAAISYY766 pKa = 6.72MTVSFVLGIVIKK778 pKa = 9.24LVCAAVTAVWALLSSIFKK796 pKa = 10.81LFGFKK801 pKa = 10.35KK802 pKa = 10.33SEE804 pKa = 4.72CEE806 pKa = 3.87FQSNDD811 pKa = 3.31GTVHH815 pKa = 4.76TVKK818 pKa = 10.59NRR820 pKa = 11.84GVEE823 pKa = 3.71IATFNSLQRR832 pKa = 11.84VNLHH836 pKa = 6.02TGIEE840 pKa = 4.38SQVGVPPTEE849 pKa = 3.93AVHH852 pKa = 6.0DD853 pKa = 3.96HH854 pKa = 6.45VYY856 pKa = 11.41ANTLLCTIDD865 pKa = 3.65GAIVGQFLGLGQDD878 pKa = 3.57VYY880 pKa = 11.18IFPKK884 pKa = 10.49HH885 pKa = 5.59FLLHH889 pKa = 5.55IRR891 pKa = 11.84KK892 pKa = 8.36MDD894 pKa = 3.95PGMVMKK900 pKa = 8.84FTSVRR905 pKa = 11.84HH906 pKa = 5.32GVTGTISCRR915 pKa = 11.84AFLQCNMQEE924 pKa = 4.15VPGYY928 pKa = 10.32DD929 pKa = 2.8IAGVSFGAVFMKK941 pKa = 10.57ANKK944 pKa = 9.91NILHH948 pKa = 6.56FFLANHH954 pKa = 5.69EE955 pKa = 4.61VKK957 pKa = 10.79SLLRR961 pKa = 11.84GNNTAVRR968 pKa = 11.84LDD970 pKa = 3.52VAHH973 pKa = 7.47PDD975 pKa = 3.31KK976 pKa = 11.05DD977 pKa = 3.8GKK979 pKa = 7.04VQRR982 pKa = 11.84YY983 pKa = 8.21IYY985 pKa = 10.22HH986 pKa = 6.34SNRR989 pKa = 11.84CEE991 pKa = 3.87YY992 pKa = 10.18VGSVTEE998 pKa = 4.06STRR1001 pKa = 11.84GDD1003 pKa = 3.64KK1004 pKa = 11.11LSGLVKK1010 pKa = 10.73YY1011 pKa = 9.47EE1012 pKa = 3.63AATVKK1017 pKa = 10.36GDD1019 pKa = 3.53CGAPLTVAEE1028 pKa = 3.86NRR1030 pKa = 11.84YY1031 pKa = 9.38YY1032 pKa = 10.98GGRR1035 pKa = 11.84CVMGLHH1041 pKa = 6.16SAGRR1045 pKa = 11.84DD1046 pKa = 3.09SAMLRR1051 pKa = 11.84EE1052 pKa = 5.16GYY1054 pKa = 8.84ATVVTQEE1061 pKa = 3.92VARR1064 pKa = 11.84EE1065 pKa = 3.97LFKK1068 pKa = 10.69TLSTYY1073 pKa = 10.81HH1074 pKa = 6.84EE1075 pKa = 5.53DD1076 pKa = 3.29IARR1079 pKa = 11.84DD1080 pKa = 4.08DD1081 pKa = 4.65IIDD1084 pKa = 3.76VPEE1087 pKa = 4.91GEE1089 pKa = 4.5TLINLQTALDD1099 pKa = 4.08AVGLTSGSFEE1109 pKa = 5.24LIGTLVEE1116 pKa = 4.17PVNISTSSKK1125 pKa = 10.39LKK1127 pKa = 10.23PSDD1130 pKa = 3.41MQRR1133 pKa = 11.84DD1134 pKa = 3.87QVFGPTPLKK1143 pKa = 10.24PAVLRR1148 pKa = 11.84TVEE1151 pKa = 3.79VDD1153 pKa = 3.51GVVQHH1158 pKa = 6.2PMVNGVKK1165 pKa = 10.09AYY1167 pKa = 8.38QTDD1170 pKa = 4.57LEE1172 pKa = 4.62CKK1174 pKa = 9.5SVRR1177 pKa = 11.84DD1178 pKa = 3.8LEE1180 pKa = 4.51LVTEE1184 pKa = 4.64IAMQKK1189 pKa = 9.14HH1190 pKa = 3.8WQATEE1195 pKa = 3.57KK1196 pKa = 10.37HH1197 pKa = 6.27SRR1199 pKa = 11.84DD1200 pKa = 3.18ILSFEE1205 pKa = 4.53DD1206 pKa = 3.88AVVPPEE1212 pKa = 3.79HH1213 pKa = 6.82WKK1215 pKa = 10.77LKK1217 pKa = 8.81PFNRR1221 pKa = 11.84KK1222 pKa = 6.73TSAGYY1227 pKa = 10.08KK1228 pKa = 8.22YY1229 pKa = 10.48RR1230 pKa = 11.84KK1231 pKa = 9.04FVTPTKK1237 pKa = 9.01PGKK1240 pKa = 10.34VFALGLEE1247 pKa = 4.23GDD1249 pKa = 3.4VDD1251 pKa = 4.41FANEE1255 pKa = 3.61NLQKK1259 pKa = 10.88VKK1261 pKa = 10.78EE1262 pKa = 4.14DD1263 pKa = 3.34TLEE1266 pKa = 4.52IISKK1270 pKa = 10.05AKK1272 pKa = 9.84EE1273 pKa = 4.21GVRR1276 pKa = 11.84TLHH1279 pKa = 6.74LCTDD1283 pKa = 4.57FLKK1286 pKa = 11.14DD1287 pKa = 3.66EE1288 pKa = 4.69LRR1290 pKa = 11.84PLAKK1294 pKa = 10.14VEE1296 pKa = 4.43SVATRR1301 pKa = 11.84VIAGTEE1307 pKa = 3.72FDD1309 pKa = 3.66YY1310 pKa = 11.47SVAVRR1315 pKa = 11.84MYY1317 pKa = 9.71FGAYY1321 pKa = 6.67MAAMFDD1327 pKa = 4.09TYY1329 pKa = 10.98VSNGMAPGINHH1340 pKa = 5.37YY1341 pKa = 10.49KK1342 pKa = 9.16EE1343 pKa = 3.73WFMLAEE1349 pKa = 4.56ALTKK1353 pKa = 10.56VGNNIFDD1360 pKa = 4.0GDD1362 pKa = 3.89FSRR1365 pKa = 11.84FDD1367 pKa = 3.34SSEE1370 pKa = 4.03QPWVHH1375 pKa = 6.44ISLLGYY1381 pKa = 9.34INRR1384 pKa = 11.84WYY1386 pKa = 10.8RR1387 pKa = 11.84FNNDD1391 pKa = 2.36KK1392 pKa = 10.65WSLEE1396 pKa = 4.06DD1397 pKa = 3.77DD1398 pKa = 3.56RR1399 pKa = 11.84VRR1401 pKa = 11.84YY1402 pKa = 9.34ILWLDD1407 pKa = 3.89LVHH1410 pKa = 6.53SRR1412 pKa = 11.84HH1413 pKa = 4.91ITGVGSRR1420 pKa = 11.84LQYY1423 pKa = 9.29VVQWNKK1429 pKa = 10.55SLPSGHH1435 pKa = 7.5PLTTIVNSMYY1445 pKa = 11.12SLVTLTGCYY1454 pKa = 9.2IAATGEE1460 pKa = 3.92LNMWDD1465 pKa = 3.56NAYY1468 pKa = 10.77LCTFGDD1474 pKa = 4.01DD1475 pKa = 3.88NVNSVSDD1482 pKa = 3.71EE1483 pKa = 3.86KK1484 pKa = 10.89RR1485 pKa = 11.84DD1486 pKa = 3.54EE1487 pKa = 4.37FNQVTVAHH1495 pKa = 6.35WMKK1498 pKa = 10.66EE1499 pKa = 4.13LFGLAYY1505 pKa = 9.17TAGHH1509 pKa = 7.13KK1510 pKa = 9.91DD1511 pKa = 3.48AQLVPYY1517 pKa = 9.76TSLDD1521 pKa = 2.98KK1522 pKa = 10.94VTFLKK1527 pKa = 10.63RR1528 pKa = 11.84SFLVDD1533 pKa = 3.8DD1534 pKa = 5.54DD1535 pKa = 4.68DD1536 pKa = 6.7AGGLIVNTPNNGWVAPLALEE1556 pKa = 4.08SFLYY1560 pKa = 9.74EE1561 pKa = 4.28GYY1563 pKa = 9.35WYY1565 pKa = 10.66KK1566 pKa = 11.0NSRR1569 pKa = 11.84DD1570 pKa = 3.44VSLDD1574 pKa = 2.96LVTRR1578 pKa = 11.84IEE1580 pKa = 4.12HH1581 pKa = 6.08SVLEE1585 pKa = 4.35MALHH1589 pKa = 5.86EE1590 pKa = 3.97QHH1592 pKa = 6.67VWDD1595 pKa = 4.43KK1596 pKa = 10.5YY1597 pKa = 10.14FPRR1600 pKa = 11.84LEE1602 pKa = 4.23EE1603 pKa = 3.72WCSRR1607 pKa = 11.84SGVCIGLKK1615 pKa = 9.92NRR1617 pKa = 11.84SAARR1621 pKa = 11.84NEE1623 pKa = 3.92VKK1625 pKa = 9.94TRR1627 pKa = 11.84FDD1629 pKa = 2.94VWFF1632 pKa = 4.41
MM1 pKa = 8.15DD2 pKa = 3.6NQINRR7 pKa = 11.84FAPPQKK13 pKa = 8.9VHH15 pKa = 6.55AVIKK19 pKa = 6.88TTVVDD24 pKa = 4.55FRR26 pKa = 11.84VQRR29 pKa = 11.84LSRR32 pKa = 11.84KK33 pKa = 9.32YY34 pKa = 10.96LDD36 pKa = 4.22GSLTNQGAKK45 pKa = 8.78QKK47 pKa = 10.54RR48 pKa = 11.84QFDD51 pKa = 4.12RR52 pKa = 11.84LSSSDD57 pKa = 3.06KK58 pKa = 10.91RR59 pKa = 11.84EE60 pKa = 3.68WRR62 pKa = 11.84EE63 pKa = 3.43ALQRR67 pKa = 11.84AIANVKK73 pKa = 8.09NHH75 pKa = 6.75CAMQALRR82 pKa = 11.84GGKK85 pKa = 9.35LRR87 pKa = 11.84GKK89 pKa = 9.06SAHH92 pKa = 6.08RR93 pKa = 11.84FEE95 pKa = 6.15RR96 pKa = 11.84MTKK99 pKa = 10.49LEE101 pKa = 4.28AALISQFDD109 pKa = 3.69EE110 pKa = 4.01RR111 pKa = 11.84VEE113 pKa = 4.24FQSGIAVAAGSVAAASIAFASYY135 pKa = 7.36QTKK138 pKa = 10.08KK139 pKa = 10.81VSDD142 pKa = 4.01SVATAVDD149 pKa = 3.37TGNDD153 pKa = 3.61LMRR156 pKa = 11.84LLIDD160 pKa = 3.57QINGFVSKK168 pKa = 9.33ITDD171 pKa = 3.33VGSVMSKK178 pKa = 9.75IALAILLVWMMVNYY192 pKa = 9.81GHH194 pKa = 7.41LPIFVAVVLACAGSYY209 pKa = 10.25LPEE212 pKa = 6.15AIEE215 pKa = 4.4MIKK218 pKa = 10.57SIAPKK223 pKa = 10.45GISLQNGGVSLAAHH237 pKa = 7.18LLSLGCTMWIPGKK250 pKa = 9.37DD251 pKa = 3.49VKK253 pKa = 11.08SVTGEE258 pKa = 3.78FLKK261 pKa = 10.66RR262 pKa = 11.84ASYY265 pKa = 9.99FPRR268 pKa = 11.84ATEE271 pKa = 4.44GIEE274 pKa = 4.25AFMKK278 pKa = 10.43KK279 pKa = 9.75GLEE282 pKa = 3.93MFEE285 pKa = 4.14GFINFVLRR293 pKa = 11.84RR294 pKa = 11.84NEE296 pKa = 3.9EE297 pKa = 3.81SWISLQGKK305 pKa = 7.42TDD307 pKa = 3.42VFVSWKK313 pKa = 9.76RR314 pKa = 11.84KK315 pKa = 8.46VIAMLKK321 pKa = 10.0FMQEE325 pKa = 3.87QPVVPIEE332 pKa = 4.5KK333 pKa = 9.58IRR335 pKa = 11.84EE336 pKa = 4.24CKK338 pKa = 10.29DD339 pKa = 3.41LLLTGYY345 pKa = 10.55GLYY348 pKa = 9.84QVLVTQEE355 pKa = 4.3SKK357 pKa = 11.06RR358 pKa = 11.84EE359 pKa = 3.78LNFWIEE365 pKa = 3.84KK366 pKa = 9.94LAVKK370 pKa = 7.65MQPHH374 pKa = 5.6QGALQAEE381 pKa = 4.89SNIRR385 pKa = 11.84PMPYY389 pKa = 9.34FIMIGGSTGVGKK401 pKa = 9.16TSLMRR406 pKa = 11.84LIGSTILMLSGEE418 pKa = 4.56VKK420 pKa = 10.07PSEE423 pKa = 4.13ALEE426 pKa = 4.2HH427 pKa = 6.47LWQKK431 pKa = 9.09GTTEE435 pKa = 3.87YY436 pKa = 10.38WNGYY440 pKa = 8.27VGQKK444 pKa = 10.04CLIMDD449 pKa = 4.68DD450 pKa = 3.53AFQVKK455 pKa = 9.56GKK457 pKa = 9.33PGDD460 pKa = 3.66MDD462 pKa = 4.04SEE464 pKa = 4.26AMQVIRR470 pKa = 11.84AVGNWSYY477 pKa = 11.29PLNFADD483 pKa = 5.86LEE485 pKa = 4.83SKK487 pKa = 10.84GKK489 pKa = 10.44FYY491 pKa = 11.26LDD493 pKa = 3.37TPLVIGTTNSRR504 pKa = 11.84NVKK507 pKa = 10.45AEE509 pKa = 3.58WAPFITEE516 pKa = 3.8PEE518 pKa = 3.83ALVRR522 pKa = 11.84RR523 pKa = 11.84FQGSYY528 pKa = 9.46WIEE531 pKa = 3.88LNPDD535 pKa = 3.17YY536 pKa = 10.07KK537 pKa = 10.93TEE539 pKa = 4.45DD540 pKa = 3.14GRR542 pKa = 11.84FDD544 pKa = 3.4FTKK547 pKa = 10.86VNDD550 pKa = 3.8EE551 pKa = 4.16FRR553 pKa = 11.84GALQRR558 pKa = 11.84IAAMKK563 pKa = 10.37SEE565 pKa = 3.72GRR567 pKa = 11.84RR568 pKa = 11.84LSVSDD573 pKa = 5.36VMDD576 pKa = 3.46QMPWQVWTLKK586 pKa = 9.78QHH588 pKa = 6.05GFDD591 pKa = 4.1RR592 pKa = 11.84EE593 pKa = 4.3NISEE597 pKa = 4.07DD598 pKa = 3.53TAPGGLRR605 pKa = 11.84NAVEE609 pKa = 3.63IAARR613 pKa = 11.84EE614 pKa = 3.7IRR616 pKa = 11.84QRR618 pKa = 11.84RR619 pKa = 11.84EE620 pKa = 3.88SNVEE624 pKa = 3.7QIQDD628 pKa = 3.76LKK630 pKa = 11.18NWTSILGEE638 pKa = 4.29ALDD641 pKa = 5.49ADD643 pKa = 4.85LEE645 pKa = 4.51LQSGLPPSMSAFVNAMSEE663 pKa = 4.2TGISDD668 pKa = 3.59TVNRR672 pKa = 11.84RR673 pKa = 11.84LFEE676 pKa = 3.87VSTRR680 pKa = 11.84QDD682 pKa = 3.46EE683 pKa = 4.43IQEE686 pKa = 4.05EE687 pKa = 4.62SQWDD691 pKa = 3.31IPMIGVQPDD700 pKa = 4.13DD701 pKa = 3.66FVGWLSSDD709 pKa = 3.68DD710 pKa = 4.44DD711 pKa = 4.7RR712 pKa = 11.84SEE714 pKa = 4.13TQSMVDD720 pKa = 3.13GVYY723 pKa = 10.42IMPEE727 pKa = 3.83AEE729 pKa = 4.27VEE731 pKa = 3.94HH732 pKa = 7.42HH733 pKa = 6.89GILRR737 pKa = 11.84KK738 pKa = 10.05LYY740 pKa = 10.44DD741 pKa = 3.75VLTSWFDD748 pKa = 3.36KK749 pKa = 11.1LPIRR753 pKa = 11.84KK754 pKa = 8.14MVAGVLAAAISYY766 pKa = 6.72MTVSFVLGIVIKK778 pKa = 9.24LVCAAVTAVWALLSSIFKK796 pKa = 10.81LFGFKK801 pKa = 10.35KK802 pKa = 10.33SEE804 pKa = 4.72CEE806 pKa = 3.87FQSNDD811 pKa = 3.31GTVHH815 pKa = 4.76TVKK818 pKa = 10.59NRR820 pKa = 11.84GVEE823 pKa = 3.71IATFNSLQRR832 pKa = 11.84VNLHH836 pKa = 6.02TGIEE840 pKa = 4.38SQVGVPPTEE849 pKa = 3.93AVHH852 pKa = 6.0DD853 pKa = 3.96HH854 pKa = 6.45VYY856 pKa = 11.41ANTLLCTIDD865 pKa = 3.65GAIVGQFLGLGQDD878 pKa = 3.57VYY880 pKa = 11.18IFPKK884 pKa = 10.49HH885 pKa = 5.59FLLHH889 pKa = 5.55IRR891 pKa = 11.84KK892 pKa = 8.36MDD894 pKa = 3.95PGMVMKK900 pKa = 8.84FTSVRR905 pKa = 11.84HH906 pKa = 5.32GVTGTISCRR915 pKa = 11.84AFLQCNMQEE924 pKa = 4.15VPGYY928 pKa = 10.32DD929 pKa = 2.8IAGVSFGAVFMKK941 pKa = 10.57ANKK944 pKa = 9.91NILHH948 pKa = 6.56FFLANHH954 pKa = 5.69EE955 pKa = 4.61VKK957 pKa = 10.79SLLRR961 pKa = 11.84GNNTAVRR968 pKa = 11.84LDD970 pKa = 3.52VAHH973 pKa = 7.47PDD975 pKa = 3.31KK976 pKa = 11.05DD977 pKa = 3.8GKK979 pKa = 7.04VQRR982 pKa = 11.84YY983 pKa = 8.21IYY985 pKa = 10.22HH986 pKa = 6.34SNRR989 pKa = 11.84CEE991 pKa = 3.87YY992 pKa = 10.18VGSVTEE998 pKa = 4.06STRR1001 pKa = 11.84GDD1003 pKa = 3.64KK1004 pKa = 11.11LSGLVKK1010 pKa = 10.73YY1011 pKa = 9.47EE1012 pKa = 3.63AATVKK1017 pKa = 10.36GDD1019 pKa = 3.53CGAPLTVAEE1028 pKa = 3.86NRR1030 pKa = 11.84YY1031 pKa = 9.38YY1032 pKa = 10.98GGRR1035 pKa = 11.84CVMGLHH1041 pKa = 6.16SAGRR1045 pKa = 11.84DD1046 pKa = 3.09SAMLRR1051 pKa = 11.84EE1052 pKa = 5.16GYY1054 pKa = 8.84ATVVTQEE1061 pKa = 3.92VARR1064 pKa = 11.84EE1065 pKa = 3.97LFKK1068 pKa = 10.69TLSTYY1073 pKa = 10.81HH1074 pKa = 6.84EE1075 pKa = 5.53DD1076 pKa = 3.29IARR1079 pKa = 11.84DD1080 pKa = 4.08DD1081 pKa = 4.65IIDD1084 pKa = 3.76VPEE1087 pKa = 4.91GEE1089 pKa = 4.5TLINLQTALDD1099 pKa = 4.08AVGLTSGSFEE1109 pKa = 5.24LIGTLVEE1116 pKa = 4.17PVNISTSSKK1125 pKa = 10.39LKK1127 pKa = 10.23PSDD1130 pKa = 3.41MQRR1133 pKa = 11.84DD1134 pKa = 3.87QVFGPTPLKK1143 pKa = 10.24PAVLRR1148 pKa = 11.84TVEE1151 pKa = 3.79VDD1153 pKa = 3.51GVVQHH1158 pKa = 6.2PMVNGVKK1165 pKa = 10.09AYY1167 pKa = 8.38QTDD1170 pKa = 4.57LEE1172 pKa = 4.62CKK1174 pKa = 9.5SVRR1177 pKa = 11.84DD1178 pKa = 3.8LEE1180 pKa = 4.51LVTEE1184 pKa = 4.64IAMQKK1189 pKa = 9.14HH1190 pKa = 3.8WQATEE1195 pKa = 3.57KK1196 pKa = 10.37HH1197 pKa = 6.27SRR1199 pKa = 11.84DD1200 pKa = 3.18ILSFEE1205 pKa = 4.53DD1206 pKa = 3.88AVVPPEE1212 pKa = 3.79HH1213 pKa = 6.82WKK1215 pKa = 10.77LKK1217 pKa = 8.81PFNRR1221 pKa = 11.84KK1222 pKa = 6.73TSAGYY1227 pKa = 10.08KK1228 pKa = 8.22YY1229 pKa = 10.48RR1230 pKa = 11.84KK1231 pKa = 9.04FVTPTKK1237 pKa = 9.01PGKK1240 pKa = 10.34VFALGLEE1247 pKa = 4.23GDD1249 pKa = 3.4VDD1251 pKa = 4.41FANEE1255 pKa = 3.61NLQKK1259 pKa = 10.88VKK1261 pKa = 10.78EE1262 pKa = 4.14DD1263 pKa = 3.34TLEE1266 pKa = 4.52IISKK1270 pKa = 10.05AKK1272 pKa = 9.84EE1273 pKa = 4.21GVRR1276 pKa = 11.84TLHH1279 pKa = 6.74LCTDD1283 pKa = 4.57FLKK1286 pKa = 11.14DD1287 pKa = 3.66EE1288 pKa = 4.69LRR1290 pKa = 11.84PLAKK1294 pKa = 10.14VEE1296 pKa = 4.43SVATRR1301 pKa = 11.84VIAGTEE1307 pKa = 3.72FDD1309 pKa = 3.66YY1310 pKa = 11.47SVAVRR1315 pKa = 11.84MYY1317 pKa = 9.71FGAYY1321 pKa = 6.67MAAMFDD1327 pKa = 4.09TYY1329 pKa = 10.98VSNGMAPGINHH1340 pKa = 5.37YY1341 pKa = 10.49KK1342 pKa = 9.16EE1343 pKa = 3.73WFMLAEE1349 pKa = 4.56ALTKK1353 pKa = 10.56VGNNIFDD1360 pKa = 4.0GDD1362 pKa = 3.89FSRR1365 pKa = 11.84FDD1367 pKa = 3.34SSEE1370 pKa = 4.03QPWVHH1375 pKa = 6.44ISLLGYY1381 pKa = 9.34INRR1384 pKa = 11.84WYY1386 pKa = 10.8RR1387 pKa = 11.84FNNDD1391 pKa = 2.36KK1392 pKa = 10.65WSLEE1396 pKa = 4.06DD1397 pKa = 3.77DD1398 pKa = 3.56RR1399 pKa = 11.84VRR1401 pKa = 11.84YY1402 pKa = 9.34ILWLDD1407 pKa = 3.89LVHH1410 pKa = 6.53SRR1412 pKa = 11.84HH1413 pKa = 4.91ITGVGSRR1420 pKa = 11.84LQYY1423 pKa = 9.29VVQWNKK1429 pKa = 10.55SLPSGHH1435 pKa = 7.5PLTTIVNSMYY1445 pKa = 11.12SLVTLTGCYY1454 pKa = 9.2IAATGEE1460 pKa = 3.92LNMWDD1465 pKa = 3.56NAYY1468 pKa = 10.77LCTFGDD1474 pKa = 4.01DD1475 pKa = 3.88NVNSVSDD1482 pKa = 3.71EE1483 pKa = 3.86KK1484 pKa = 10.89RR1485 pKa = 11.84DD1486 pKa = 3.54EE1487 pKa = 4.37FNQVTVAHH1495 pKa = 6.35WMKK1498 pKa = 10.66EE1499 pKa = 4.13LFGLAYY1505 pKa = 9.17TAGHH1509 pKa = 7.13KK1510 pKa = 9.91DD1511 pKa = 3.48AQLVPYY1517 pKa = 9.76TSLDD1521 pKa = 2.98KK1522 pKa = 10.94VTFLKK1527 pKa = 10.63RR1528 pKa = 11.84SFLVDD1533 pKa = 3.8DD1534 pKa = 5.54DD1535 pKa = 4.68DD1536 pKa = 6.7AGGLIVNTPNNGWVAPLALEE1556 pKa = 4.08SFLYY1560 pKa = 9.74EE1561 pKa = 4.28GYY1563 pKa = 9.35WYY1565 pKa = 10.66KK1566 pKa = 11.0NSRR1569 pKa = 11.84DD1570 pKa = 3.44VSLDD1574 pKa = 2.96LVTRR1578 pKa = 11.84IEE1580 pKa = 4.12HH1581 pKa = 6.08SVLEE1585 pKa = 4.35MALHH1589 pKa = 5.86EE1590 pKa = 3.97QHH1592 pKa = 6.67VWDD1595 pKa = 4.43KK1596 pKa = 10.5YY1597 pKa = 10.14FPRR1600 pKa = 11.84LEE1602 pKa = 4.23EE1603 pKa = 3.72WCSRR1607 pKa = 11.84SGVCIGLKK1615 pKa = 9.92NRR1617 pKa = 11.84SAARR1621 pKa = 11.84NEE1623 pKa = 3.92VKK1625 pKa = 9.94TRR1627 pKa = 11.84FDD1629 pKa = 2.94VWFF1632 pKa = 4.41
Molecular weight: 183.33 kDa
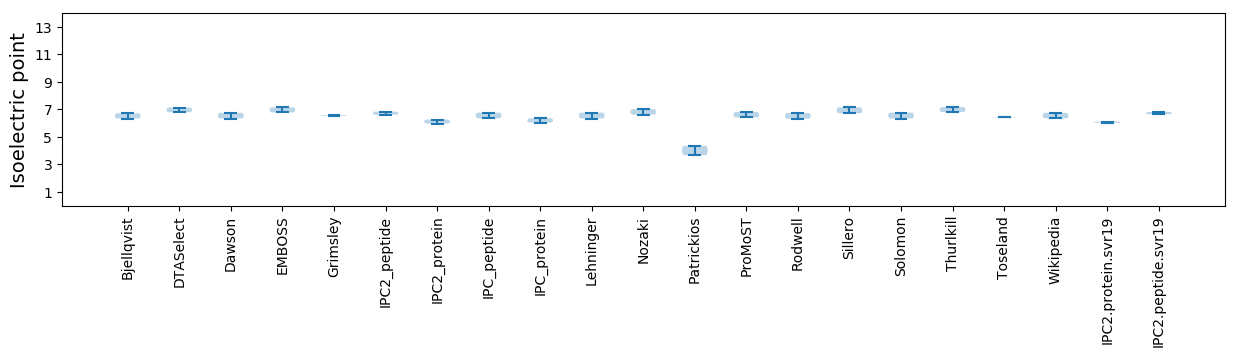
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KJC4|A0A1L3KJC4_9VIRU Uncharacterized protein OS=Sanxia picorna-like virus 11 OX=1923368 PE=4 SV=1
MM1 pKa = 7.85DD2 pKa = 5.31FDD4 pKa = 3.49STAAFRR10 pKa = 11.84TFLSPVNWDD19 pKa = 3.56RR20 pKa = 11.84LKK22 pKa = 11.09GAVGVRR28 pKa = 11.84GTLKK32 pKa = 10.26FHH34 pKa = 6.59LVVAATPFHH43 pKa = 6.38QGLAYY48 pKa = 9.83MCFQYY53 pKa = 10.69GISVADD59 pKa = 4.16GNWNRR64 pKa = 11.84GNVYY68 pKa = 10.73NLATNLPHH76 pKa = 6.2VRR78 pKa = 11.84LNIAEE83 pKa = 4.23EE84 pKa = 4.31TMATLSVPFVSNRR97 pKa = 11.84EE98 pKa = 4.14YY99 pKa = 10.03ITVDD103 pKa = 3.04NNVADD108 pKa = 3.56NTLNYY113 pKa = 10.84GVFNLGLLLGSPVVAGQTPPSYY135 pKa = 10.58SLYY138 pKa = 11.02VSFHH142 pKa = 6.44DD143 pKa = 4.99LEE145 pKa = 6.0LIGAVPYY152 pKa = 8.68NTTNVVLQTGVVIKK166 pKa = 10.76DD167 pKa = 3.52SVSLGKK173 pKa = 10.56SGVTAEE179 pKa = 4.19ANAGGVISKK188 pKa = 8.5TLEE191 pKa = 3.72VAADD195 pKa = 3.58VAKK198 pKa = 10.08AGSMIPSLSKK208 pKa = 10.65IGGTADD214 pKa = 2.57WMLRR218 pKa = 11.84SLSKK222 pKa = 9.48TASSFGFSKK231 pKa = 11.01PNDD234 pKa = 3.37EE235 pKa = 4.56TKK237 pKa = 6.98WTKK240 pKa = 7.81MVKK243 pKa = 9.91YY244 pKa = 9.33GYY246 pKa = 10.36AGEE249 pKa = 4.4SQVDD253 pKa = 3.94VPNQAFPLSPFMSNKK268 pKa = 9.6IAVTSAIGCVDD279 pKa = 4.01EE280 pKa = 5.74DD281 pKa = 4.03QMSFDD286 pKa = 3.97NVLTKK291 pKa = 10.02PSYY294 pKa = 9.99IFRR297 pKa = 11.84GQFSDD302 pKa = 3.5AAASGDD308 pKa = 3.8LLYY311 pKa = 11.07GCSLSPSCFWYY322 pKa = 10.4RR323 pKa = 11.84DD324 pKa = 3.59FSVVGSPNGNIGLPTNATVTQNCFYY349 pKa = 10.4PSTLCYY355 pKa = 10.24IGDD358 pKa = 4.05NFRR361 pKa = 11.84MWRR364 pKa = 11.84GSFKK368 pKa = 10.69FRR370 pKa = 11.84ITFAKK375 pKa = 9.47TKK377 pKa = 9.63LHH379 pKa = 6.16GGRR382 pKa = 11.84VQVSYY387 pKa = 11.3VPYY390 pKa = 8.85SQAVSAIAAPSNTVAAPEE408 pKa = 4.72IITSLVQPSGYY419 pKa = 10.18SEE421 pKa = 4.45IFDD424 pKa = 4.81LKK426 pKa = 10.85DD427 pKa = 3.28SSIVEE432 pKa = 4.58FVCPYY437 pKa = 10.2ISPDD441 pKa = 3.27PYY443 pKa = 10.56IAFNNAMGSLTMIVVNPLRR462 pKa = 11.84VTGNAPTAVQFMVEE476 pKa = 4.07VCAEE480 pKa = 4.14PGFEE484 pKa = 4.29LSVPKK489 pKa = 10.14PSLLHH494 pKa = 5.65PVGKK498 pKa = 10.41GGLVAPKK505 pKa = 9.01YY506 pKa = 10.11QSGLEE511 pKa = 4.17VPGPRR516 pKa = 11.84IPLVSNLDD524 pKa = 3.5STSQFVIGEE533 pKa = 4.33KK534 pKa = 10.53FNSLKK539 pKa = 10.35QLMMMPDD546 pKa = 3.11WSYY549 pKa = 11.79FEE551 pKa = 5.26APNASFTDD559 pKa = 4.38FVPVPWFKK567 pKa = 10.98TNTVTNTTPVSNTYY581 pKa = 9.78SVSWFGSRR589 pKa = 11.84SSRR592 pKa = 11.84VSQLFAFANGSTAYY606 pKa = 9.36TIHH609 pKa = 7.31HH610 pKa = 7.48DD611 pKa = 3.63GANVPGITVTMRR623 pKa = 11.84WWGNNNGVIGGITDD637 pKa = 4.14LRR639 pKa = 11.84APEE642 pKa = 4.31NNSFSSVFIPEE653 pKa = 3.88EE654 pKa = 3.73QATSRR659 pKa = 11.84FIVPTYY665 pKa = 9.77SKK667 pKa = 11.2YY668 pKa = 9.82MRR670 pKa = 11.84IPIMRR675 pKa = 11.84STPAFGTNVTSLFDD689 pKa = 3.6PMTYY693 pKa = 9.31TGEE696 pKa = 4.09YY697 pKa = 9.12SLAMPLCVLRR707 pKa = 11.84NTSGASRR714 pKa = 11.84RR715 pKa = 11.84FVVGRR720 pKa = 11.84AAADD724 pKa = 3.38DD725 pKa = 4.62AYY727 pKa = 10.66ASQFVGPPPCVLFPTTSTNSPVNGGFSSLNQYY759 pKa = 9.9
MM1 pKa = 7.85DD2 pKa = 5.31FDD4 pKa = 3.49STAAFRR10 pKa = 11.84TFLSPVNWDD19 pKa = 3.56RR20 pKa = 11.84LKK22 pKa = 11.09GAVGVRR28 pKa = 11.84GTLKK32 pKa = 10.26FHH34 pKa = 6.59LVVAATPFHH43 pKa = 6.38QGLAYY48 pKa = 9.83MCFQYY53 pKa = 10.69GISVADD59 pKa = 4.16GNWNRR64 pKa = 11.84GNVYY68 pKa = 10.73NLATNLPHH76 pKa = 6.2VRR78 pKa = 11.84LNIAEE83 pKa = 4.23EE84 pKa = 4.31TMATLSVPFVSNRR97 pKa = 11.84EE98 pKa = 4.14YY99 pKa = 10.03ITVDD103 pKa = 3.04NNVADD108 pKa = 3.56NTLNYY113 pKa = 10.84GVFNLGLLLGSPVVAGQTPPSYY135 pKa = 10.58SLYY138 pKa = 11.02VSFHH142 pKa = 6.44DD143 pKa = 4.99LEE145 pKa = 6.0LIGAVPYY152 pKa = 8.68NTTNVVLQTGVVIKK166 pKa = 10.76DD167 pKa = 3.52SVSLGKK173 pKa = 10.56SGVTAEE179 pKa = 4.19ANAGGVISKK188 pKa = 8.5TLEE191 pKa = 3.72VAADD195 pKa = 3.58VAKK198 pKa = 10.08AGSMIPSLSKK208 pKa = 10.65IGGTADD214 pKa = 2.57WMLRR218 pKa = 11.84SLSKK222 pKa = 9.48TASSFGFSKK231 pKa = 11.01PNDD234 pKa = 3.37EE235 pKa = 4.56TKK237 pKa = 6.98WTKK240 pKa = 7.81MVKK243 pKa = 9.91YY244 pKa = 9.33GYY246 pKa = 10.36AGEE249 pKa = 4.4SQVDD253 pKa = 3.94VPNQAFPLSPFMSNKK268 pKa = 9.6IAVTSAIGCVDD279 pKa = 4.01EE280 pKa = 5.74DD281 pKa = 4.03QMSFDD286 pKa = 3.97NVLTKK291 pKa = 10.02PSYY294 pKa = 9.99IFRR297 pKa = 11.84GQFSDD302 pKa = 3.5AAASGDD308 pKa = 3.8LLYY311 pKa = 11.07GCSLSPSCFWYY322 pKa = 10.4RR323 pKa = 11.84DD324 pKa = 3.59FSVVGSPNGNIGLPTNATVTQNCFYY349 pKa = 10.4PSTLCYY355 pKa = 10.24IGDD358 pKa = 4.05NFRR361 pKa = 11.84MWRR364 pKa = 11.84GSFKK368 pKa = 10.69FRR370 pKa = 11.84ITFAKK375 pKa = 9.47TKK377 pKa = 9.63LHH379 pKa = 6.16GGRR382 pKa = 11.84VQVSYY387 pKa = 11.3VPYY390 pKa = 8.85SQAVSAIAAPSNTVAAPEE408 pKa = 4.72IITSLVQPSGYY419 pKa = 10.18SEE421 pKa = 4.45IFDD424 pKa = 4.81LKK426 pKa = 10.85DD427 pKa = 3.28SSIVEE432 pKa = 4.58FVCPYY437 pKa = 10.2ISPDD441 pKa = 3.27PYY443 pKa = 10.56IAFNNAMGSLTMIVVNPLRR462 pKa = 11.84VTGNAPTAVQFMVEE476 pKa = 4.07VCAEE480 pKa = 4.14PGFEE484 pKa = 4.29LSVPKK489 pKa = 10.14PSLLHH494 pKa = 5.65PVGKK498 pKa = 10.41GGLVAPKK505 pKa = 9.01YY506 pKa = 10.11QSGLEE511 pKa = 4.17VPGPRR516 pKa = 11.84IPLVSNLDD524 pKa = 3.5STSQFVIGEE533 pKa = 4.33KK534 pKa = 10.53FNSLKK539 pKa = 10.35QLMMMPDD546 pKa = 3.11WSYY549 pKa = 11.79FEE551 pKa = 5.26APNASFTDD559 pKa = 4.38FVPVPWFKK567 pKa = 10.98TNTVTNTTPVSNTYY581 pKa = 9.78SVSWFGSRR589 pKa = 11.84SSRR592 pKa = 11.84VSQLFAFANGSTAYY606 pKa = 9.36TIHH609 pKa = 7.31HH610 pKa = 7.48DD611 pKa = 3.63GANVPGITVTMRR623 pKa = 11.84WWGNNNGVIGGITDD637 pKa = 4.14LRR639 pKa = 11.84APEE642 pKa = 4.31NNSFSSVFIPEE653 pKa = 3.88EE654 pKa = 3.73QATSRR659 pKa = 11.84FIVPTYY665 pKa = 9.77SKK667 pKa = 11.2YY668 pKa = 9.82MRR670 pKa = 11.84IPIMRR675 pKa = 11.84STPAFGTNVTSLFDD689 pKa = 3.6PMTYY693 pKa = 9.31TGEE696 pKa = 4.09YY697 pKa = 9.12SLAMPLCVLRR707 pKa = 11.84NTSGASRR714 pKa = 11.84RR715 pKa = 11.84FVVGRR720 pKa = 11.84AAADD724 pKa = 3.38DD725 pKa = 4.62AYY727 pKa = 10.66ASQFVGPPPCVLFPTTSTNSPVNGGFSSLNQYY759 pKa = 9.9
Molecular weight: 82.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2391 |
759 |
1632 |
1195.5 |
132.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.445 ± 0.154 | 1.213 ± 0.049 |
5.228 ± 0.598 | 5.186 ± 1.072 |
4.977 ± 0.508 | 7.277 ± 0.233 |
1.924 ± 0.408 | 4.81 ± 0.278 |
5.102 ± 0.848 | 8.281 ± 0.67 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.928 ± 0.137 | 4.726 ± 0.749 |
4.517 ± 1.156 | 3.43 ± 0.372 |
4.726 ± 0.61 | 7.946 ± 1.154 |
6.023 ± 0.635 | 9.118 ± 0.235 |
1.84 ± 0.183 | 3.304 ± 0.304 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
