
Nocardiopsis dassonvillei (strain ATCC 23218 / DSM 43111 / CIP 107115 / JCM 7437 / KCTC 9190 / NBRC 14626 / NCTC 10488 / NRRL B-5397 / IMRU 509) (Actinomadura dassonvillei)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptosporangiales; Nocardiopsaceae; Nocardiopsis; Nocardiopsis dassonvillei; Nocardiopsis dassonvillei subsp. dassonvillei
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins
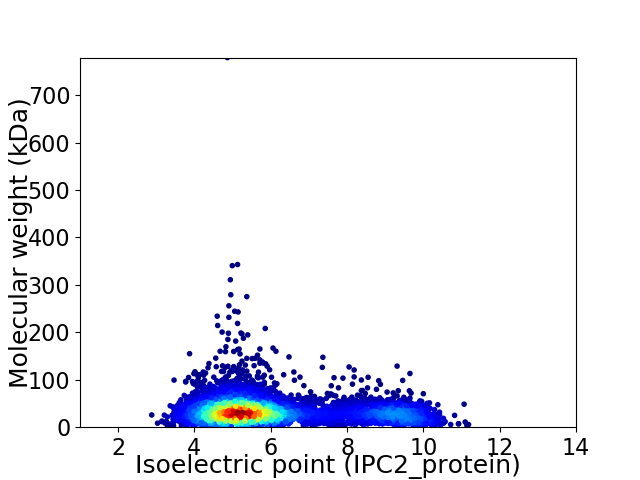
Virtual 2D-PAGE plot for 5497 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D7AYD5|D7AYD5_NOCDD Transcriptional regulator TetR family OS=Nocardiopsis dassonvillei (strain ATCC 23218 / DSM 43111 / CIP 107115 / JCM 7437 / KCTC 9190 / NBRC 14626 / NCTC 10488 / NRRL B-5397 / IMRU 509) OX=446468 GN=Ndas_0675 PE=4 SV=1
MM1 pKa = 7.59RR2 pKa = 11.84VRR4 pKa = 11.84RR5 pKa = 11.84ISTILGGTAALAFALSACGTADD27 pKa = 3.09VGGGSGGEE35 pKa = 4.12GGGGEE40 pKa = 4.39GGGDD44 pKa = 3.72TITIGVKK51 pKa = 9.95YY52 pKa = 9.32DD53 pKa = 3.47QPGLGLLDD61 pKa = 3.69GDD63 pKa = 4.28APVGFDD69 pKa = 3.63VDD71 pKa = 3.77VATYY75 pKa = 9.9IAGEE79 pKa = 4.01LGYY82 pKa = 10.55EE83 pKa = 4.15PEE85 pKa = 4.14QIEE88 pKa = 4.21WTEE91 pKa = 3.56AASANRR97 pKa = 11.84EE98 pKa = 4.07SFLQQGTVDD107 pKa = 4.09MVIATYY113 pKa = 10.53SITDD117 pKa = 3.23EE118 pKa = 3.92RR119 pKa = 11.84RR120 pKa = 11.84QVVDD124 pKa = 3.69FAGPYY129 pKa = 9.6YY130 pKa = 10.51VAQQDD135 pKa = 3.78ILVQADD141 pKa = 3.49NEE143 pKa = 4.74DD144 pKa = 3.42IQGPEE149 pKa = 3.85DD150 pKa = 3.42LAGKK154 pKa = 7.48VLCSASGSRR163 pKa = 11.84SANNIVDD170 pKa = 3.96DD171 pKa = 4.0MGIDD175 pKa = 3.43AEE177 pKa = 4.2LRR179 pKa = 11.84EE180 pKa = 4.33AGNYY184 pKa = 8.72SEE186 pKa = 5.36CLSLLGNGTVDD197 pKa = 5.55AVTTDD202 pKa = 3.32NTILAGFAAQDD213 pKa = 3.46PGTYY217 pKa = 8.65RR218 pKa = 11.84VVNNPFGEE226 pKa = 3.99EE227 pKa = 4.2RR228 pKa = 11.84YY229 pKa = 10.31GVGLPAGSTEE239 pKa = 3.79RR240 pKa = 11.84CEE242 pKa = 4.25EE243 pKa = 3.92VNAAIQKK250 pKa = 8.31MWDD253 pKa = 2.84EE254 pKa = 4.94GYY256 pKa = 10.88AVEE259 pKa = 4.79ALEE262 pKa = 4.29TAFGEE267 pKa = 4.04TDD269 pKa = 3.54FAYY272 pKa = 10.2EE273 pKa = 3.85EE274 pKa = 4.19TAPEE278 pKa = 3.79VDD280 pKa = 3.19ACQQ283 pKa = 3.25
MM1 pKa = 7.59RR2 pKa = 11.84VRR4 pKa = 11.84RR5 pKa = 11.84ISTILGGTAALAFALSACGTADD27 pKa = 3.09VGGGSGGEE35 pKa = 4.12GGGGEE40 pKa = 4.39GGGDD44 pKa = 3.72TITIGVKK51 pKa = 9.95YY52 pKa = 9.32DD53 pKa = 3.47QPGLGLLDD61 pKa = 3.69GDD63 pKa = 4.28APVGFDD69 pKa = 3.63VDD71 pKa = 3.77VATYY75 pKa = 9.9IAGEE79 pKa = 4.01LGYY82 pKa = 10.55EE83 pKa = 4.15PEE85 pKa = 4.14QIEE88 pKa = 4.21WTEE91 pKa = 3.56AASANRR97 pKa = 11.84EE98 pKa = 4.07SFLQQGTVDD107 pKa = 4.09MVIATYY113 pKa = 10.53SITDD117 pKa = 3.23EE118 pKa = 3.92RR119 pKa = 11.84RR120 pKa = 11.84QVVDD124 pKa = 3.69FAGPYY129 pKa = 9.6YY130 pKa = 10.51VAQQDD135 pKa = 3.78ILVQADD141 pKa = 3.49NEE143 pKa = 4.74DD144 pKa = 3.42IQGPEE149 pKa = 3.85DD150 pKa = 3.42LAGKK154 pKa = 7.48VLCSASGSRR163 pKa = 11.84SANNIVDD170 pKa = 3.96DD171 pKa = 4.0MGIDD175 pKa = 3.43AEE177 pKa = 4.2LRR179 pKa = 11.84EE180 pKa = 4.33AGNYY184 pKa = 8.72SEE186 pKa = 5.36CLSLLGNGTVDD197 pKa = 5.55AVTTDD202 pKa = 3.32NTILAGFAAQDD213 pKa = 3.46PGTYY217 pKa = 8.65RR218 pKa = 11.84VVNNPFGEE226 pKa = 3.99EE227 pKa = 4.2RR228 pKa = 11.84YY229 pKa = 10.31GVGLPAGSTEE239 pKa = 3.79RR240 pKa = 11.84CEE242 pKa = 4.25EE243 pKa = 3.92VNAAIQKK250 pKa = 8.31MWDD253 pKa = 2.84EE254 pKa = 4.94GYY256 pKa = 10.88AVEE259 pKa = 4.79ALEE262 pKa = 4.29TAFGEE267 pKa = 4.04TDD269 pKa = 3.54FAYY272 pKa = 10.2EE273 pKa = 3.85EE274 pKa = 4.19TAPEE278 pKa = 3.79VDD280 pKa = 3.19ACQQ283 pKa = 3.25
Molecular weight: 29.56 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D7AX38|D7AX38_NOCDD Non-specific serine/threonine protein kinase OS=Nocardiopsis dassonvillei (strain ATCC 23218 / DSM 43111 / CIP 107115 / JCM 7437 / KCTC 9190 / NBRC 14626 / NCTC 10488 / NRRL B-5397 / IMRU 509) OX=446468 GN=Ndas_4419 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.72HH17 pKa = 5.74RR18 pKa = 11.84KK19 pKa = 7.77LLKK22 pKa = 7.46RR23 pKa = 11.84TRR25 pKa = 11.84VARR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.94KK32 pKa = 9.77
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.72HH17 pKa = 5.74RR18 pKa = 11.84KK19 pKa = 7.77LLKK22 pKa = 7.46RR23 pKa = 11.84TRR25 pKa = 11.84VARR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.94KK32 pKa = 9.77
Molecular weight: 3.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1834850 |
30 |
7422 |
333.8 |
35.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.107 ± 0.051 | 0.705 ± 0.01 |
6.158 ± 0.03 | 6.542 ± 0.032 |
2.747 ± 0.018 | 9.798 ± 0.034 |
2.277 ± 0.017 | 2.804 ± 0.026 |
1.396 ± 0.02 | 10.275 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.845 ± 0.015 | 1.721 ± 0.017 |
6.191 ± 0.033 | 2.445 ± 0.02 |
8.538 ± 0.037 | 5.256 ± 0.026 |
5.752 ± 0.024 | 8.988 ± 0.038 |
1.494 ± 0.014 | 1.962 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
