
Microbulbifer agarilyticus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Cellvibrionales; Microbulbiferaceae; Microbulbifer
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
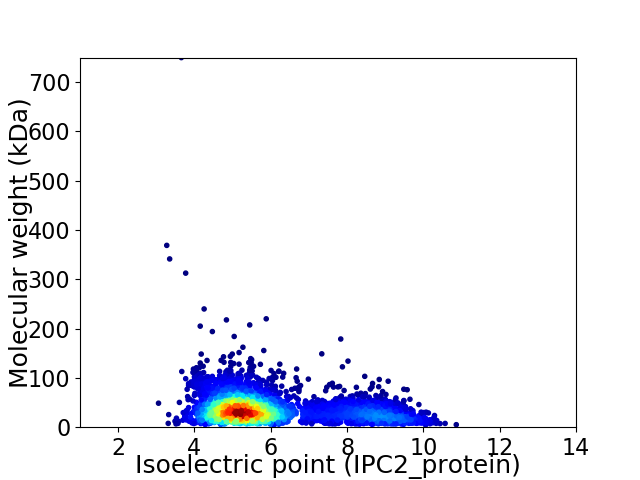
Virtual 2D-PAGE plot for 3454 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Q2M9N2|A0A1Q2M9N2_9GAMM VWFA domain-containing protein OS=Microbulbifer agarilyticus OX=260552 GN=Mag101_05340 PE=4 SV=1
MM1 pKa = 6.78EE2 pKa = 4.28TSFAIIEE9 pKa = 4.01VDD11 pKa = 3.61EE12 pKa = 4.51AATAYY17 pKa = 8.29EE18 pKa = 3.91QLVTINDD25 pKa = 3.73YY26 pKa = 11.77AEE28 pKa = 4.49VPVAWSKK35 pKa = 10.63WSGDD39 pKa = 3.53PATTAQYY46 pKa = 11.19LLNGQVVLEE55 pKa = 4.07QNVGGGATQTGTATLQVAEE74 pKa = 4.64GGQYY78 pKa = 10.47SLQVALCNDD87 pKa = 4.41DD88 pKa = 4.0GCSTSAATDD97 pKa = 3.26IVVADD102 pKa = 4.05TDD104 pKa = 4.39GSHH107 pKa = 7.45LDD109 pKa = 4.54PITVTAGEE117 pKa = 4.14NNKK120 pKa = 10.15PYY122 pKa = 10.37TNSTNSVVGTYY133 pKa = 8.99FVEE136 pKa = 3.68WGVYY140 pKa = 7.95GRR142 pKa = 11.84KK143 pKa = 9.56FSVDD147 pKa = 3.16MMPSYY152 pKa = 11.49NLTHH156 pKa = 7.4LIYY159 pKa = 10.65GFIPICGGDD168 pKa = 4.17GINDD172 pKa = 3.6SLKK175 pKa = 10.38EE176 pKa = 4.02IEE178 pKa = 5.01GSFQALQRR186 pKa = 11.84SCSGRR191 pKa = 11.84EE192 pKa = 3.78DD193 pKa = 3.97FKK195 pKa = 11.81VSLHH199 pKa = 6.9DD200 pKa = 4.64PFAALQKK207 pKa = 10.36SQADD211 pKa = 3.6QTFSDD216 pKa = 4.67PYY218 pKa = 10.38KK219 pKa = 11.14GNFGQLMALKK229 pKa = 9.89QAYY232 pKa = 7.94PDD234 pKa = 4.18LKK236 pKa = 10.08ILPSIGGWTLSDD248 pKa = 3.3PFYY251 pKa = 10.79FFSDD255 pKa = 3.0AAKK258 pKa = 10.23RR259 pKa = 11.84KK260 pKa = 7.43TFVDD264 pKa = 3.65SVEE267 pKa = 3.9EE268 pKa = 5.27FIRR271 pKa = 11.84TWKK274 pKa = 10.55FFDD277 pKa = 4.25GVDD280 pKa = 3.86IDD282 pKa = 3.61WEE284 pKa = 4.58YY285 pKa = 11.03PGGQGANPTLGDD297 pKa = 3.6PAIDD301 pKa = 3.78GEE303 pKa = 4.65TYY305 pKa = 10.53RR306 pKa = 11.84LLMRR310 pKa = 11.84DD311 pKa = 3.73LRR313 pKa = 11.84AMLDD317 pKa = 3.43NLEE320 pKa = 4.19QEE322 pKa = 4.27TGRR325 pKa = 11.84TYY327 pKa = 11.39EE328 pKa = 3.95LTSAIGAGSDD338 pKa = 3.35KK339 pKa = 11.1IEE341 pKa = 4.43DD342 pKa = 3.22VDD344 pKa = 4.04YY345 pKa = 11.67LDD347 pKa = 3.46VQQYY351 pKa = 8.23MDD353 pKa = 3.88YY354 pKa = 10.1FFVMTYY360 pKa = 10.69DD361 pKa = 5.02FYY363 pKa = 11.64GGWSNEE369 pKa = 4.11VLGHH373 pKa = 5.33QAALYY378 pKa = 9.93APAWKK383 pKa = 9.54PDD385 pKa = 3.36TDD387 pKa = 3.91YY388 pKa = 7.94TTHH391 pKa = 7.19NGIQNLLGLGVDD403 pKa = 3.51PGKK406 pKa = 10.7LVVGAAMYY414 pKa = 10.31GRR416 pKa = 11.84GWKK419 pKa = 9.8GVSGWTGNDD428 pKa = 2.9HH429 pKa = 5.69MTGTATGMVNGTWEE443 pKa = 4.75DD444 pKa = 3.8GVVDD448 pKa = 3.56YY449 pKa = 11.27RR450 pKa = 11.84DD451 pKa = 3.3IVSRR455 pKa = 11.84IATGEE460 pKa = 3.68WEE462 pKa = 4.35EE463 pKa = 4.98YY464 pKa = 10.27YY465 pKa = 10.94DD466 pKa = 3.96STAEE470 pKa = 3.55APYY473 pKa = 10.12IFKK476 pKa = 10.55PSTGDD481 pKa = 3.68LITYY485 pKa = 9.37DD486 pKa = 3.45DD487 pKa = 4.72HH488 pKa = 8.32RR489 pKa = 11.84SVMAKK494 pKa = 9.81GAYY497 pKa = 8.43VQSNNLAGLFSWEE510 pKa = 3.53IDD512 pKa = 3.3ADD514 pKa = 3.73NGDD517 pKa = 4.0IMNAMHH523 pKa = 7.08EE524 pKa = 4.59SLGHH528 pKa = 6.47GDD530 pKa = 3.46GFGNRR535 pKa = 11.84APVARR540 pKa = 11.84AGGDD544 pKa = 3.29QSVDD548 pKa = 3.06SGASVSLDD556 pKa = 2.81GSTSSDD562 pKa = 3.59LDD564 pKa = 3.84NDD566 pKa = 3.99PLTYY570 pKa = 10.51SWVQVSGTSVTLQNASSASASFTAPAVSADD600 pKa = 3.68EE601 pKa = 4.21EE602 pKa = 4.97LVFSLTVSDD611 pKa = 5.51GEE613 pKa = 4.85ASDD616 pKa = 4.02SDD618 pKa = 3.94QVSVTVVADD627 pKa = 3.81QPNQAPSADD636 pKa = 3.41AGADD640 pKa = 3.54QMVTTPATVTLNGSASSDD658 pKa = 3.57PDD660 pKa = 3.54GDD662 pKa = 3.82ALTYY666 pKa = 10.76SWVQVAGSSVTLSDD680 pKa = 3.97ASSATPSFSAAEE692 pKa = 4.04VSAEE696 pKa = 3.85QEE698 pKa = 4.16LVFEE702 pKa = 5.27LNVNDD707 pKa = 5.19GSLSDD712 pKa = 3.66ATPDD716 pKa = 3.3QVSIFLLPADD726 pKa = 4.78ANTPPQVSAPAQVTIQEE743 pKa = 4.46GASDD747 pKa = 4.76SITATGTDD755 pKa = 3.32ADD757 pKa = 4.5GDD759 pKa = 4.13ALTYY763 pKa = 8.45TWSGMVSGSGDD774 pKa = 3.98TISITAPQVEE784 pKa = 4.59ADD786 pKa = 3.84TNFTLTVTVSDD797 pKa = 5.52GIASASADD805 pKa = 3.31VTVTVTNVIVDD816 pKa = 4.25GGCSSTDD823 pKa = 3.37PNAGNVPAWQSGSTYY838 pKa = 10.67LGGDD842 pKa = 3.51QVSHH846 pKa = 6.2EE847 pKa = 4.04QLVWQAKK854 pKa = 8.35YY855 pKa = 7.14WTQQEE860 pKa = 4.49PGFSSADD867 pKa = 3.2WQLVSDD873 pKa = 5.78IEE875 pKa = 4.63VPWNASTAYY884 pKa = 10.63NGGDD888 pKa = 3.43EE889 pKa = 4.39VNHH892 pKa = 5.64EE893 pKa = 4.39GKK895 pKa = 10.49RR896 pKa = 11.84YY897 pKa = 8.33RR898 pKa = 11.84AKK900 pKa = 9.79WWTQGQNPSSSSDD913 pKa = 2.93WEE915 pKa = 4.37EE916 pKa = 5.01IGDD919 pKa = 3.92ASCNN923 pKa = 3.64
MM1 pKa = 6.78EE2 pKa = 4.28TSFAIIEE9 pKa = 4.01VDD11 pKa = 3.61EE12 pKa = 4.51AATAYY17 pKa = 8.29EE18 pKa = 3.91QLVTINDD25 pKa = 3.73YY26 pKa = 11.77AEE28 pKa = 4.49VPVAWSKK35 pKa = 10.63WSGDD39 pKa = 3.53PATTAQYY46 pKa = 11.19LLNGQVVLEE55 pKa = 4.07QNVGGGATQTGTATLQVAEE74 pKa = 4.64GGQYY78 pKa = 10.47SLQVALCNDD87 pKa = 4.41DD88 pKa = 4.0GCSTSAATDD97 pKa = 3.26IVVADD102 pKa = 4.05TDD104 pKa = 4.39GSHH107 pKa = 7.45LDD109 pKa = 4.54PITVTAGEE117 pKa = 4.14NNKK120 pKa = 10.15PYY122 pKa = 10.37TNSTNSVVGTYY133 pKa = 8.99FVEE136 pKa = 3.68WGVYY140 pKa = 7.95GRR142 pKa = 11.84KK143 pKa = 9.56FSVDD147 pKa = 3.16MMPSYY152 pKa = 11.49NLTHH156 pKa = 7.4LIYY159 pKa = 10.65GFIPICGGDD168 pKa = 4.17GINDD172 pKa = 3.6SLKK175 pKa = 10.38EE176 pKa = 4.02IEE178 pKa = 5.01GSFQALQRR186 pKa = 11.84SCSGRR191 pKa = 11.84EE192 pKa = 3.78DD193 pKa = 3.97FKK195 pKa = 11.81VSLHH199 pKa = 6.9DD200 pKa = 4.64PFAALQKK207 pKa = 10.36SQADD211 pKa = 3.6QTFSDD216 pKa = 4.67PYY218 pKa = 10.38KK219 pKa = 11.14GNFGQLMALKK229 pKa = 9.89QAYY232 pKa = 7.94PDD234 pKa = 4.18LKK236 pKa = 10.08ILPSIGGWTLSDD248 pKa = 3.3PFYY251 pKa = 10.79FFSDD255 pKa = 3.0AAKK258 pKa = 10.23RR259 pKa = 11.84KK260 pKa = 7.43TFVDD264 pKa = 3.65SVEE267 pKa = 3.9EE268 pKa = 5.27FIRR271 pKa = 11.84TWKK274 pKa = 10.55FFDD277 pKa = 4.25GVDD280 pKa = 3.86IDD282 pKa = 3.61WEE284 pKa = 4.58YY285 pKa = 11.03PGGQGANPTLGDD297 pKa = 3.6PAIDD301 pKa = 3.78GEE303 pKa = 4.65TYY305 pKa = 10.53RR306 pKa = 11.84LLMRR310 pKa = 11.84DD311 pKa = 3.73LRR313 pKa = 11.84AMLDD317 pKa = 3.43NLEE320 pKa = 4.19QEE322 pKa = 4.27TGRR325 pKa = 11.84TYY327 pKa = 11.39EE328 pKa = 3.95LTSAIGAGSDD338 pKa = 3.35KK339 pKa = 11.1IEE341 pKa = 4.43DD342 pKa = 3.22VDD344 pKa = 4.04YY345 pKa = 11.67LDD347 pKa = 3.46VQQYY351 pKa = 8.23MDD353 pKa = 3.88YY354 pKa = 10.1FFVMTYY360 pKa = 10.69DD361 pKa = 5.02FYY363 pKa = 11.64GGWSNEE369 pKa = 4.11VLGHH373 pKa = 5.33QAALYY378 pKa = 9.93APAWKK383 pKa = 9.54PDD385 pKa = 3.36TDD387 pKa = 3.91YY388 pKa = 7.94TTHH391 pKa = 7.19NGIQNLLGLGVDD403 pKa = 3.51PGKK406 pKa = 10.7LVVGAAMYY414 pKa = 10.31GRR416 pKa = 11.84GWKK419 pKa = 9.8GVSGWTGNDD428 pKa = 2.9HH429 pKa = 5.69MTGTATGMVNGTWEE443 pKa = 4.75DD444 pKa = 3.8GVVDD448 pKa = 3.56YY449 pKa = 11.27RR450 pKa = 11.84DD451 pKa = 3.3IVSRR455 pKa = 11.84IATGEE460 pKa = 3.68WEE462 pKa = 4.35EE463 pKa = 4.98YY464 pKa = 10.27YY465 pKa = 10.94DD466 pKa = 3.96STAEE470 pKa = 3.55APYY473 pKa = 10.12IFKK476 pKa = 10.55PSTGDD481 pKa = 3.68LITYY485 pKa = 9.37DD486 pKa = 3.45DD487 pKa = 4.72HH488 pKa = 8.32RR489 pKa = 11.84SVMAKK494 pKa = 9.81GAYY497 pKa = 8.43VQSNNLAGLFSWEE510 pKa = 3.53IDD512 pKa = 3.3ADD514 pKa = 3.73NGDD517 pKa = 4.0IMNAMHH523 pKa = 7.08EE524 pKa = 4.59SLGHH528 pKa = 6.47GDD530 pKa = 3.46GFGNRR535 pKa = 11.84APVARR540 pKa = 11.84AGGDD544 pKa = 3.29QSVDD548 pKa = 3.06SGASVSLDD556 pKa = 2.81GSTSSDD562 pKa = 3.59LDD564 pKa = 3.84NDD566 pKa = 3.99PLTYY570 pKa = 10.51SWVQVSGTSVTLQNASSASASFTAPAVSADD600 pKa = 3.68EE601 pKa = 4.21EE602 pKa = 4.97LVFSLTVSDD611 pKa = 5.51GEE613 pKa = 4.85ASDD616 pKa = 4.02SDD618 pKa = 3.94QVSVTVVADD627 pKa = 3.81QPNQAPSADD636 pKa = 3.41AGADD640 pKa = 3.54QMVTTPATVTLNGSASSDD658 pKa = 3.57PDD660 pKa = 3.54GDD662 pKa = 3.82ALTYY666 pKa = 10.76SWVQVAGSSVTLSDD680 pKa = 3.97ASSATPSFSAAEE692 pKa = 4.04VSAEE696 pKa = 3.85QEE698 pKa = 4.16LVFEE702 pKa = 5.27LNVNDD707 pKa = 5.19GSLSDD712 pKa = 3.66ATPDD716 pKa = 3.3QVSIFLLPADD726 pKa = 4.78ANTPPQVSAPAQVTIQEE743 pKa = 4.46GASDD747 pKa = 4.76SITATGTDD755 pKa = 3.32ADD757 pKa = 4.5GDD759 pKa = 4.13ALTYY763 pKa = 8.45TWSGMVSGSGDD774 pKa = 3.98TISITAPQVEE784 pKa = 4.59ADD786 pKa = 3.84TNFTLTVTVSDD797 pKa = 5.52GIASASADD805 pKa = 3.31VTVTVTNVIVDD816 pKa = 4.25GGCSSTDD823 pKa = 3.37PNAGNVPAWQSGSTYY838 pKa = 10.67LGGDD842 pKa = 3.51QVSHH846 pKa = 6.2EE847 pKa = 4.04QLVWQAKK854 pKa = 8.35YY855 pKa = 7.14WTQQEE860 pKa = 4.49PGFSSADD867 pKa = 3.2WQLVSDD873 pKa = 5.78IEE875 pKa = 4.63VPWNASTAYY884 pKa = 10.63NGGDD888 pKa = 3.43EE889 pKa = 4.39VNHH892 pKa = 5.64EE893 pKa = 4.39GKK895 pKa = 10.49RR896 pKa = 11.84YY897 pKa = 8.33RR898 pKa = 11.84AKK900 pKa = 9.79WWTQGQNPSSSSDD913 pKa = 2.93WEE915 pKa = 4.37EE916 pKa = 5.01IGDD919 pKa = 3.92ASCNN923 pKa = 3.64
Molecular weight: 98.31 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Q2M9B5|A0A1Q2M9B5_9GAMM GntR family transcriptional regulator OS=Microbulbifer agarilyticus OX=260552 GN=Mag101_01875 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.08RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.92GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.2SGRR28 pKa = 11.84KK29 pKa = 8.27ILARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.95GRR39 pKa = 11.84KK40 pKa = 8.19VLCAA44 pKa = 3.83
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLKK11 pKa = 10.08RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.92GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.2SGRR28 pKa = 11.84KK29 pKa = 8.27ILARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.95GRR39 pKa = 11.84KK40 pKa = 8.19VLCAA44 pKa = 3.83
Molecular weight: 5.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1229174 |
38 |
7326 |
355.9 |
39.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.199 ± 0.052 | 0.976 ± 0.014 |
5.857 ± 0.047 | 6.455 ± 0.041 |
3.893 ± 0.027 | 7.939 ± 0.045 |
2.101 ± 0.022 | 5.095 ± 0.026 |
3.879 ± 0.039 | 10.223 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.306 ± 0.02 | 3.636 ± 0.032 |
4.53 ± 0.038 | 4.266 ± 0.032 |
5.786 ± 0.053 | 6.313 ± 0.047 |
5.129 ± 0.039 | 7.124 ± 0.038 |
1.421 ± 0.017 | 2.874 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
