
Bradyrhizobium sp. LMTR 3
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Bradyrhizobiaceae; Bradyrhizobium; unclassified Bradyrhizobium
Average proteome isoelectric point is 6.97
Get precalculated fractions of proteins
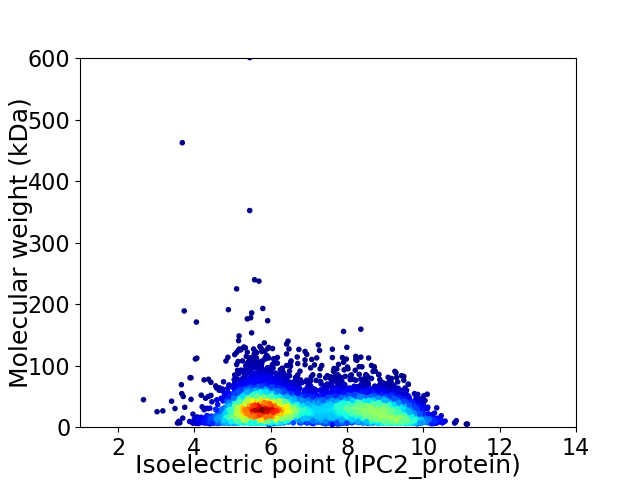
Virtual 2D-PAGE plot for 6979 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B9YI77|A0A1B9YI77_9BRAD Uncharacterized protein OS=Bradyrhizobium sp. LMTR 3 OX=189873 GN=LMTR3_26510 PE=4 SV=1
MM1 pKa = 7.6ASASFVEE8 pKa = 5.22IISHH12 pKa = 6.11FAGYY16 pKa = 7.91MQIFEE21 pKa = 5.46DD22 pKa = 3.51IARR25 pKa = 11.84DD26 pKa = 3.81RR27 pKa = 11.84IQYY30 pKa = 10.68DD31 pKa = 3.65EE32 pKa = 4.53TPGPGPSGDD41 pKa = 3.72YY42 pKa = 7.55TTLRR46 pKa = 11.84PNYY49 pKa = 7.8HH50 pKa = 6.31HH51 pKa = 7.59RR52 pKa = 11.84YY53 pKa = 9.44APEE56 pKa = 5.37DD57 pKa = 3.7MDD59 pKa = 4.46SAASPVPEE67 pKa = 4.89VIADD71 pKa = 3.94DD72 pKa = 4.36PMDD75 pKa = 4.52MIRR78 pKa = 11.84GRR80 pKa = 11.84PLKK83 pKa = 10.16LLKK86 pKa = 10.6GPQDD90 pKa = 3.38PDD92 pKa = 3.32FDD94 pKa = 4.83FFPTPPKK101 pKa = 10.22PNIVLPPGGGGGGGGGSEE119 pKa = 3.86FNIRR123 pKa = 11.84VKK125 pKa = 11.23YY126 pKa = 9.83EE127 pKa = 4.07DD128 pKa = 3.98GGAEE132 pKa = 3.79IQLTVHH138 pKa = 5.31QHH140 pKa = 3.86NFMRR144 pKa = 11.84DD145 pKa = 2.87EE146 pKa = 4.63DD147 pKa = 4.58FNLPADD153 pKa = 4.06FLVAAEE159 pKa = 4.21PLIMSLNSDD168 pKa = 3.06AMATIEE174 pKa = 4.1QLVANASAEE183 pKa = 4.07IPTDD187 pKa = 1.98WWMPQNDD194 pKa = 3.63AGATDD199 pKa = 4.2FLITHH204 pKa = 7.05DD205 pKa = 4.53AAWADD210 pKa = 3.84RR211 pKa = 11.84DD212 pKa = 4.01GTPDD216 pKa = 3.32EE217 pKa = 4.8HH218 pKa = 7.28SVPAGYY224 pKa = 9.91YY225 pKa = 10.41VNGEE229 pKa = 4.02LQEE232 pKa = 4.7PPTEE236 pKa = 3.94PRR238 pKa = 11.84PPIEE242 pKa = 4.85LKK244 pKa = 10.44EE245 pKa = 4.17LPDD248 pKa = 3.66TGDD251 pKa = 5.66GIGQWASMGGNYY263 pKa = 10.09SINAALLVDD272 pKa = 3.71IGEE275 pKa = 4.56SARR278 pKa = 11.84TMVVLGDD285 pKa = 3.84YY286 pKa = 10.13FKK288 pKa = 10.81TDD290 pKa = 4.33AMFQTNTTVDD300 pKa = 3.44NDD302 pKa = 3.7EE303 pKa = 4.33VSVSGGEE310 pKa = 4.11GTPSVTSEE318 pKa = 4.08GNVATNIANFVQNPSIYY335 pKa = 10.71ADD337 pKa = 3.98LPSYY341 pKa = 9.26WAGPNWIVDD350 pKa = 4.03VVDD353 pKa = 3.42GDD355 pKa = 4.29YY356 pKa = 11.54YY357 pKa = 11.46SVNTVSQTNYY367 pKa = 10.65LSDD370 pKa = 3.78NDD372 pKa = 3.5VATQVSSEE380 pKa = 3.66GHH382 pKa = 5.73YY383 pKa = 11.1NLVSGHH389 pKa = 5.57NQLGNLAQIFDD400 pKa = 4.28GEE402 pKa = 4.1IHH404 pKa = 6.77YY405 pKa = 10.67DD406 pKa = 3.86LIIIEE411 pKa = 4.29GAYY414 pKa = 10.44HH415 pKa = 6.21GMNVIFQNNILLNNDD430 pKa = 4.32NIVMSADD437 pKa = 3.33GTDD440 pKa = 3.56PSQSVNSGHH449 pKa = 6.67NSLLNEE455 pKa = 4.23GAIEE459 pKa = 4.19NYY461 pKa = 10.64GGDD464 pKa = 3.79TFDD467 pKa = 4.38GLTPGLQQILDD478 pKa = 4.06LLASGVTSIDD488 pKa = 3.58PEE490 pKa = 4.58LGSIIAGNGGPLRR503 pKa = 11.84VLYY506 pKa = 10.9VKK508 pKa = 10.74GDD510 pKa = 3.7YY511 pKa = 11.02YY512 pKa = 11.0DD513 pKa = 5.09INAVWQTNITSDD525 pKa = 3.92INVLYY530 pKa = 10.27QLQNEE535 pKa = 4.49PSPDD539 pKa = 3.78LMALDD544 pKa = 4.59PDD546 pKa = 4.23ADD548 pKa = 4.11VTQSASTGGNEE559 pKa = 4.48LANDD563 pKa = 3.85AAIVDD568 pKa = 4.0VNPDD572 pKa = 3.36VIYY575 pKa = 11.25VEE577 pKa = 4.66GEE579 pKa = 4.27VYY581 pKa = 10.17TDD583 pKa = 4.2SILVQANLLPVDD595 pKa = 3.76QDD597 pKa = 3.73DD598 pKa = 4.69AVNGDD603 pKa = 3.39TDD605 pKa = 3.87TLVTEE610 pKa = 5.62LIAFVDD616 pKa = 3.96DD617 pKa = 4.38TQDD620 pKa = 3.18QTNASPAVVTNSVQADD636 pKa = 3.57PMASVLHH643 pKa = 6.19
MM1 pKa = 7.6ASASFVEE8 pKa = 5.22IISHH12 pKa = 6.11FAGYY16 pKa = 7.91MQIFEE21 pKa = 5.46DD22 pKa = 3.51IARR25 pKa = 11.84DD26 pKa = 3.81RR27 pKa = 11.84IQYY30 pKa = 10.68DD31 pKa = 3.65EE32 pKa = 4.53TPGPGPSGDD41 pKa = 3.72YY42 pKa = 7.55TTLRR46 pKa = 11.84PNYY49 pKa = 7.8HH50 pKa = 6.31HH51 pKa = 7.59RR52 pKa = 11.84YY53 pKa = 9.44APEE56 pKa = 5.37DD57 pKa = 3.7MDD59 pKa = 4.46SAASPVPEE67 pKa = 4.89VIADD71 pKa = 3.94DD72 pKa = 4.36PMDD75 pKa = 4.52MIRR78 pKa = 11.84GRR80 pKa = 11.84PLKK83 pKa = 10.16LLKK86 pKa = 10.6GPQDD90 pKa = 3.38PDD92 pKa = 3.32FDD94 pKa = 4.83FFPTPPKK101 pKa = 10.22PNIVLPPGGGGGGGGGSEE119 pKa = 3.86FNIRR123 pKa = 11.84VKK125 pKa = 11.23YY126 pKa = 9.83EE127 pKa = 4.07DD128 pKa = 3.98GGAEE132 pKa = 3.79IQLTVHH138 pKa = 5.31QHH140 pKa = 3.86NFMRR144 pKa = 11.84DD145 pKa = 2.87EE146 pKa = 4.63DD147 pKa = 4.58FNLPADD153 pKa = 4.06FLVAAEE159 pKa = 4.21PLIMSLNSDD168 pKa = 3.06AMATIEE174 pKa = 4.1QLVANASAEE183 pKa = 4.07IPTDD187 pKa = 1.98WWMPQNDD194 pKa = 3.63AGATDD199 pKa = 4.2FLITHH204 pKa = 7.05DD205 pKa = 4.53AAWADD210 pKa = 3.84RR211 pKa = 11.84DD212 pKa = 4.01GTPDD216 pKa = 3.32EE217 pKa = 4.8HH218 pKa = 7.28SVPAGYY224 pKa = 9.91YY225 pKa = 10.41VNGEE229 pKa = 4.02LQEE232 pKa = 4.7PPTEE236 pKa = 3.94PRR238 pKa = 11.84PPIEE242 pKa = 4.85LKK244 pKa = 10.44EE245 pKa = 4.17LPDD248 pKa = 3.66TGDD251 pKa = 5.66GIGQWASMGGNYY263 pKa = 10.09SINAALLVDD272 pKa = 3.71IGEE275 pKa = 4.56SARR278 pKa = 11.84TMVVLGDD285 pKa = 3.84YY286 pKa = 10.13FKK288 pKa = 10.81TDD290 pKa = 4.33AMFQTNTTVDD300 pKa = 3.44NDD302 pKa = 3.7EE303 pKa = 4.33VSVSGGEE310 pKa = 4.11GTPSVTSEE318 pKa = 4.08GNVATNIANFVQNPSIYY335 pKa = 10.71ADD337 pKa = 3.98LPSYY341 pKa = 9.26WAGPNWIVDD350 pKa = 4.03VVDD353 pKa = 3.42GDD355 pKa = 4.29YY356 pKa = 11.54YY357 pKa = 11.46SVNTVSQTNYY367 pKa = 10.65LSDD370 pKa = 3.78NDD372 pKa = 3.5VATQVSSEE380 pKa = 3.66GHH382 pKa = 5.73YY383 pKa = 11.1NLVSGHH389 pKa = 5.57NQLGNLAQIFDD400 pKa = 4.28GEE402 pKa = 4.1IHH404 pKa = 6.77YY405 pKa = 10.67DD406 pKa = 3.86LIIIEE411 pKa = 4.29GAYY414 pKa = 10.44HH415 pKa = 6.21GMNVIFQNNILLNNDD430 pKa = 4.32NIVMSADD437 pKa = 3.33GTDD440 pKa = 3.56PSQSVNSGHH449 pKa = 6.67NSLLNEE455 pKa = 4.23GAIEE459 pKa = 4.19NYY461 pKa = 10.64GGDD464 pKa = 3.79TFDD467 pKa = 4.38GLTPGLQQILDD478 pKa = 4.06LLASGVTSIDD488 pKa = 3.58PEE490 pKa = 4.58LGSIIAGNGGPLRR503 pKa = 11.84VLYY506 pKa = 10.9VKK508 pKa = 10.74GDD510 pKa = 3.7YY511 pKa = 11.02YY512 pKa = 11.0DD513 pKa = 5.09INAVWQTNITSDD525 pKa = 3.92INVLYY530 pKa = 10.27QLQNEE535 pKa = 4.49PSPDD539 pKa = 3.78LMALDD544 pKa = 4.59PDD546 pKa = 4.23ADD548 pKa = 4.11VTQSASTGGNEE559 pKa = 4.48LANDD563 pKa = 3.85AAIVDD568 pKa = 4.0VNPDD572 pKa = 3.36VIYY575 pKa = 11.25VEE577 pKa = 4.66GEE579 pKa = 4.27VYY581 pKa = 10.17TDD583 pKa = 4.2SILVQANLLPVDD595 pKa = 3.76QDD597 pKa = 3.73DD598 pKa = 4.69AVNGDD603 pKa = 3.39TDD605 pKa = 3.87TLVTEE610 pKa = 5.62LIAFVDD616 pKa = 3.96DD617 pKa = 4.38TQDD620 pKa = 3.18QTNASPAVVTNSVQADD636 pKa = 3.57PMASVLHH643 pKa = 6.19
Molecular weight: 69.1 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B9Z3M2|A0A1B9Z3M2_9BRAD Probable GTP-binding protein EngB OS=Bradyrhizobium sp. LMTR 3 OX=189873 GN=engB PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.55GFRR19 pKa = 11.84ARR21 pKa = 11.84LATAGGRR28 pKa = 11.84KK29 pKa = 8.95VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.55GFRR19 pKa = 11.84ARR21 pKa = 11.84LATAGGRR28 pKa = 11.84KK29 pKa = 8.95VLAARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.03RR41 pKa = 11.84LSAA44 pKa = 4.03
Molecular weight: 5.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2128772 |
29 |
5480 |
305.0 |
33.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.364 ± 0.044 | 0.88 ± 0.01 |
5.437 ± 0.026 | 5.428 ± 0.028 |
3.779 ± 0.021 | 8.297 ± 0.032 |
2.038 ± 0.015 | 5.429 ± 0.021 |
3.74 ± 0.029 | 9.869 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.489 ± 0.015 | 2.791 ± 0.02 |
5.25 ± 0.026 | 3.175 ± 0.016 |
7.008 ± 0.033 | 5.65 ± 0.022 |
5.312 ± 0.027 | 7.456 ± 0.025 |
1.339 ± 0.014 | 2.269 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
