
Heliothis virescens (Tobacco budworm moth)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Panarthropoda; Arthropoda; Mandibulata; Pancrustacea; Hexapoda; Insecta; Dicondylia; Pterygota; Neoptera; Endopterygota; Amphiesmenoptera; Lepidoptera; Glossata; Neolepidoptera;
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
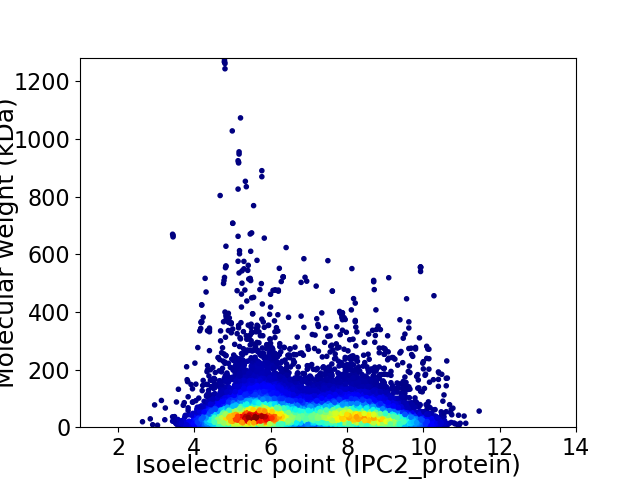
Virtual 2D-PAGE plot for 17055 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2A4JLX9|A0A2A4JLX9_HELVI Uncharacterized protein OS=Heliothis virescens OX=7102 GN=B5V51_853 PE=3 SV=1
LL1 pKa = 7.98SPIVALVPTTSSTTTTTKK19 pKa = 10.65KK20 pKa = 8.99PTTACPHH27 pKa = 6.21ATTKK31 pKa = 9.47PTVKK35 pKa = 8.14TTTQKK40 pKa = 9.4PTTTKK45 pKa = 10.28TSTTICSKK53 pKa = 7.51TTTRR57 pKa = 11.84KK58 pKa = 9.23PVATTPTEE66 pKa = 3.87PDD68 pKa = 2.58ITTTKK73 pKa = 9.98TYY75 pKa = 8.55HH76 pKa = 4.97TTPNVDD82 pKa = 2.37ITTPGLNTITPIEE95 pKa = 4.02PHH97 pKa = 5.33VTTPSGPDD105 pKa = 3.06VTTPTGPEE113 pKa = 3.75VTTPGGPEE121 pKa = 3.68VTTPGGPEE129 pKa = 3.68VTTPGGPEE137 pKa = 3.29ITTPGGPEE145 pKa = 3.27ITTPGGPEE153 pKa = 3.27ITTPGGPEE161 pKa = 3.68VTTPGGPEE169 pKa = 3.71LTTPSKK175 pKa = 11.16PEE177 pKa = 3.59VTTPSGPEE185 pKa = 3.24ITTPGGPEE193 pKa = 3.68VTTPGGPEE201 pKa = 3.71VTTTSGPEE209 pKa = 3.31ITTPGGPEE217 pKa = 3.69VTTPSGPEE225 pKa = 3.24ITTPGGPEE233 pKa = 3.27ITTPGGPEE241 pKa = 3.27ITTPGGPEE249 pKa = 3.68VTTPGGPEE257 pKa = 3.93LTTSSGPEE265 pKa = 3.54VTTPNGPEE273 pKa = 3.43ITTPGGPEE281 pKa = 3.69VTTPSGPEE289 pKa = 3.24ITTPGGPEE297 pKa = 3.72EE298 pKa = 4.34TTPGGPDD305 pKa = 3.08TTTAPCGHH313 pKa = 7.46DD314 pKa = 3.06EE315 pKa = 4.35TTPSEE320 pKa = 4.37TDD322 pKa = 3.23TTPAALDD329 pKa = 3.51TTTPSGPVVTTPSGPEE345 pKa = 3.7TTTAPCGHH353 pKa = 7.46DD354 pKa = 3.06EE355 pKa = 4.35TTPSEE360 pKa = 4.37TDD362 pKa = 3.23TTPAALDD369 pKa = 3.51TTTPSGPVVTTPSGPEE385 pKa = 3.7TTTAPCGHH393 pKa = 7.46DD394 pKa = 3.06EE395 pKa = 4.35TTPSEE400 pKa = 4.37TDD402 pKa = 3.23TTPAALDD409 pKa = 3.51TTTPSGPVVTTPSGPEE425 pKa = 3.7TTTAPCGHH433 pKa = 7.46DD434 pKa = 3.06EE435 pKa = 4.35TTPSEE440 pKa = 4.37TDD442 pKa = 3.23TTPAALDD449 pKa = 3.51TTTPSGPVVTTPSGPEE465 pKa = 3.7TTTAPCGHH473 pKa = 7.46DD474 pKa = 3.06EE475 pKa = 4.35TTPSEE480 pKa = 4.37TDD482 pKa = 3.23TTPAALDD489 pKa = 3.51TTTPSGPVVTTPSGPEE505 pKa = 3.7TTTAPCGHH513 pKa = 7.46DD514 pKa = 3.06EE515 pKa = 4.35TTPSEE520 pKa = 4.37TDD522 pKa = 3.23TTPAALDD529 pKa = 3.51TTTPSGPVVTTPSGPEE545 pKa = 3.7TTTAPCGHH553 pKa = 7.46DD554 pKa = 3.06EE555 pKa = 4.35TTPSEE560 pKa = 4.37TDD562 pKa = 3.23TTPAALDD569 pKa = 3.51TTTPSGPVVTTPSGPEE585 pKa = 3.7TTTAPCGHH593 pKa = 7.46DD594 pKa = 3.06EE595 pKa = 4.35TTPSEE600 pKa = 4.37TDD602 pKa = 3.23TTPAALDD609 pKa = 3.51TTTPSGPVVTTPSGPEE625 pKa = 3.7TTTAPCGHH633 pKa = 7.46DD634 pKa = 3.06EE635 pKa = 4.35TTPSEE640 pKa = 4.37TDD642 pKa = 3.23TTPAALDD649 pKa = 3.51TTTPSGPVVTTPSGPEE665 pKa = 3.7TTTAPCGHH673 pKa = 7.46DD674 pKa = 3.06EE675 pKa = 4.35TTPSEE680 pKa = 4.37TDD682 pKa = 3.23TTPAALDD689 pKa = 3.51TTTPSGPVVTTPSGPEE705 pKa = 3.7TTTAPCGHH713 pKa = 7.46DD714 pKa = 3.06EE715 pKa = 4.35TTPSEE720 pKa = 4.37TDD722 pKa = 3.23TTPAALDD729 pKa = 3.51TTTPSGPVVTTPSGPEE745 pKa = 3.7TTTAPCGHH753 pKa = 7.46DD754 pKa = 3.06EE755 pKa = 4.35TTPSEE760 pKa = 4.37TDD762 pKa = 3.23TTPAALDD769 pKa = 3.51TTTPSGPVVTTPSGPEE785 pKa = 3.7TTTAPCGHH793 pKa = 7.46DD794 pKa = 3.06EE795 pKa = 4.35TTPSEE800 pKa = 4.37TDD802 pKa = 3.23TTPAALDD809 pKa = 3.51TTTPSGPVVTTPSGPEE825 pKa = 3.7TTTAPCGHH833 pKa = 7.46DD834 pKa = 3.06EE835 pKa = 4.35TTPSEE840 pKa = 4.37TDD842 pKa = 3.23TTPAALDD849 pKa = 3.51TTTPSGPVVTTPSGPEE865 pKa = 3.7TTTAPCGHH873 pKa = 7.46DD874 pKa = 3.06EE875 pKa = 4.35TTPSEE880 pKa = 4.37TDD882 pKa = 3.3TTPAAPDD889 pKa = 3.38TTTPIGPEE897 pKa = 3.31ITTPGGPEE905 pKa = 3.68VTTPGGPEE913 pKa = 3.29ITTPGGPEE921 pKa = 3.27ITTPGGPEE929 pKa = 3.27ITTPGGPEE937 pKa = 3.68VTTPGGPEE945 pKa = 3.68VTTPGGPEE953 pKa = 3.69VTTPSGPEE961 pKa = 3.24ITTPGGPEE969 pKa = 3.27ITTPGGPEE977 pKa = 3.68VTTPGGPEE985 pKa = 3.29ITTPGGPEE993 pKa = 3.46ITTSGGPEE1001 pKa = 3.43ITTPGGPEE1009 pKa = 3.58KK1010 pKa = 8.62TTPSGPEE1017 pKa = 3.68TTTAPCGHH1025 pKa = 7.46DD1026 pKa = 3.06EE1027 pKa = 4.35TTPSEE1032 pKa = 4.37TDD1034 pKa = 3.23TTPAALDD1041 pKa = 3.51TTTPSGPVVTTPSGPEE1057 pKa = 3.7TTTAPCGHH1065 pKa = 7.46DD1066 pKa = 3.06EE1067 pKa = 4.35TTPSEE1072 pKa = 4.37TDD1074 pKa = 3.23TTPAALDD1081 pKa = 3.47TTTPSGSVVTTPSGPEE1097 pKa = 3.7TTTAPCGHH1105 pKa = 7.46DD1106 pKa = 3.06EE1107 pKa = 4.35TTPSEE1112 pKa = 4.37TDD1114 pKa = 3.23TTPAALDD1121 pKa = 3.47TTTPSGSVVTTPSGPEE1137 pKa = 3.7TTTAPCGHH1145 pKa = 7.46DD1146 pKa = 3.06EE1147 pKa = 4.35TTPSEE1152 pKa = 4.37TDD1154 pKa = 3.23TTPAALDD1161 pKa = 3.49TTTPSGLVVTTPSGPEE1177 pKa = 3.7TTTAPCGHH1185 pKa = 7.46DD1186 pKa = 3.06EE1187 pKa = 4.35TTPSEE1192 pKa = 4.37TDD1194 pKa = 3.3TTPAAPDD1201 pKa = 3.41TTTPSGPVVTTPSGPEE1217 pKa = 3.7TTTAPCGHH1225 pKa = 7.46DD1226 pKa = 3.06EE1227 pKa = 4.35TTPSEE1232 pKa = 4.44TGTTPAAPDD1241 pKa = 3.01ITTPSGPEE1249 pKa = 3.69TTTAPCGHH1257 pKa = 7.46DD1258 pKa = 3.06EE1259 pKa = 4.35TTPSEE1264 pKa = 4.44TGTTPAAPDD1273 pKa = 3.45TTTPSGPVVTTPSGPEE1289 pKa = 3.7TTTAPCGHH1297 pKa = 7.46DD1298 pKa = 3.06EE1299 pKa = 4.35TTPSEE1304 pKa = 4.37TDD1306 pKa = 3.3TTPAAPDD1313 pKa = 3.41TTTPSGPVVTTPSGPEE1329 pKa = 3.7TTTAPCGHH1337 pKa = 7.46DD1338 pKa = 3.06EE1339 pKa = 4.35TTPSEE1344 pKa = 4.44TGTTPAAPDD1353 pKa = 3.45TTTPSGPEE1361 pKa = 3.77TTTAPCGHH1369 pKa = 7.46DD1370 pKa = 3.06EE1371 pKa = 4.35TTPSEE1376 pKa = 4.44TGTTPAAPDD1385 pKa = 3.45TTTPSGPVVTTPSGPEE1401 pKa = 3.7TTTAPCGHH1409 pKa = 7.46DD1410 pKa = 3.06EE1411 pKa = 4.35TTPSEE1416 pKa = 4.37TDD1418 pKa = 3.3TTPAAPDD1425 pKa = 3.41TTTPSGPVVTTPSGPEE1441 pKa = 3.7TTTAPCGHH1449 pKa = 7.46DD1450 pKa = 3.06EE1451 pKa = 4.35TTPSEE1456 pKa = 4.37TDD1458 pKa = 3.3TTPAAPDD1465 pKa = 3.33TATPSGPEE1473 pKa = 4.12TTTSPCGHH1481 pKa = 7.45DD1482 pKa = 3.12EE1483 pKa = 4.31TTPSEE1488 pKa = 4.37TDD1490 pKa = 3.23TTPAALDD1497 pKa = 3.49TTTPSGLVVTTPSGPEE1513 pKa = 3.7TTTAPCGHH1521 pKa = 7.49DD1522 pKa = 3.29EE1523 pKa = 4.29TTPSGPHH1530 pKa = 5.08VTTQSEE1536 pKa = 4.66PDD1538 pKa = 3.35TTTPRR1543 pKa = 11.84PATFPPITTPSSCQHH1558 pKa = 6.49HH1559 pKa = 6.47PTSEE1563 pKa = 4.07KK1564 pKa = 10.57PITSTSRR1571 pKa = 11.84PSTPPCGHH1579 pKa = 6.59SSTTKK1584 pKa = 8.97ATTIHH1589 pKa = 7.24RR1590 pKa = 11.84SGKK1593 pKa = 8.83PCKK1596 pKa = 10.08CSDD1599 pKa = 3.24GATTEE1604 pKa = 4.16VPTDD1608 pKa = 3.67VSDD1611 pKa = 3.94HH1612 pKa = 6.32LVDD1615 pKa = 3.84EE1616 pKa = 4.89TVHH1619 pKa = 6.38LLNTGVVAVEE1629 pKa = 4.35DD1630 pKa = 3.99LLEE1633 pKa = 4.23EE1634 pKa = 4.37LLASGQKK1641 pKa = 10.53DD1642 pKa = 3.0ILHH1645 pKa = 7.2KK1646 pKa = 10.24IRR1648 pKa = 11.84RR1649 pKa = 11.84TKK1651 pKa = 10.34RR1652 pKa = 11.84KK1653 pKa = 8.36NRR1655 pKa = 11.84RR1656 pKa = 11.84TRR1658 pKa = 11.84LHH1660 pKa = 6.51KK1661 pKa = 10.06PATVLPILII1670 pKa = 4.64
LL1 pKa = 7.98SPIVALVPTTSSTTTTTKK19 pKa = 10.65KK20 pKa = 8.99PTTACPHH27 pKa = 6.21ATTKK31 pKa = 9.47PTVKK35 pKa = 8.14TTTQKK40 pKa = 9.4PTTTKK45 pKa = 10.28TSTTICSKK53 pKa = 7.51TTTRR57 pKa = 11.84KK58 pKa = 9.23PVATTPTEE66 pKa = 3.87PDD68 pKa = 2.58ITTTKK73 pKa = 9.98TYY75 pKa = 8.55HH76 pKa = 4.97TTPNVDD82 pKa = 2.37ITTPGLNTITPIEE95 pKa = 4.02PHH97 pKa = 5.33VTTPSGPDD105 pKa = 3.06VTTPTGPEE113 pKa = 3.75VTTPGGPEE121 pKa = 3.68VTTPGGPEE129 pKa = 3.68VTTPGGPEE137 pKa = 3.29ITTPGGPEE145 pKa = 3.27ITTPGGPEE153 pKa = 3.27ITTPGGPEE161 pKa = 3.68VTTPGGPEE169 pKa = 3.71LTTPSKK175 pKa = 11.16PEE177 pKa = 3.59VTTPSGPEE185 pKa = 3.24ITTPGGPEE193 pKa = 3.68VTTPGGPEE201 pKa = 3.71VTTTSGPEE209 pKa = 3.31ITTPGGPEE217 pKa = 3.69VTTPSGPEE225 pKa = 3.24ITTPGGPEE233 pKa = 3.27ITTPGGPEE241 pKa = 3.27ITTPGGPEE249 pKa = 3.68VTTPGGPEE257 pKa = 3.93LTTSSGPEE265 pKa = 3.54VTTPNGPEE273 pKa = 3.43ITTPGGPEE281 pKa = 3.69VTTPSGPEE289 pKa = 3.24ITTPGGPEE297 pKa = 3.72EE298 pKa = 4.34TTPGGPDD305 pKa = 3.08TTTAPCGHH313 pKa = 7.46DD314 pKa = 3.06EE315 pKa = 4.35TTPSEE320 pKa = 4.37TDD322 pKa = 3.23TTPAALDD329 pKa = 3.51TTTPSGPVVTTPSGPEE345 pKa = 3.7TTTAPCGHH353 pKa = 7.46DD354 pKa = 3.06EE355 pKa = 4.35TTPSEE360 pKa = 4.37TDD362 pKa = 3.23TTPAALDD369 pKa = 3.51TTTPSGPVVTTPSGPEE385 pKa = 3.7TTTAPCGHH393 pKa = 7.46DD394 pKa = 3.06EE395 pKa = 4.35TTPSEE400 pKa = 4.37TDD402 pKa = 3.23TTPAALDD409 pKa = 3.51TTTPSGPVVTTPSGPEE425 pKa = 3.7TTTAPCGHH433 pKa = 7.46DD434 pKa = 3.06EE435 pKa = 4.35TTPSEE440 pKa = 4.37TDD442 pKa = 3.23TTPAALDD449 pKa = 3.51TTTPSGPVVTTPSGPEE465 pKa = 3.7TTTAPCGHH473 pKa = 7.46DD474 pKa = 3.06EE475 pKa = 4.35TTPSEE480 pKa = 4.37TDD482 pKa = 3.23TTPAALDD489 pKa = 3.51TTTPSGPVVTTPSGPEE505 pKa = 3.7TTTAPCGHH513 pKa = 7.46DD514 pKa = 3.06EE515 pKa = 4.35TTPSEE520 pKa = 4.37TDD522 pKa = 3.23TTPAALDD529 pKa = 3.51TTTPSGPVVTTPSGPEE545 pKa = 3.7TTTAPCGHH553 pKa = 7.46DD554 pKa = 3.06EE555 pKa = 4.35TTPSEE560 pKa = 4.37TDD562 pKa = 3.23TTPAALDD569 pKa = 3.51TTTPSGPVVTTPSGPEE585 pKa = 3.7TTTAPCGHH593 pKa = 7.46DD594 pKa = 3.06EE595 pKa = 4.35TTPSEE600 pKa = 4.37TDD602 pKa = 3.23TTPAALDD609 pKa = 3.51TTTPSGPVVTTPSGPEE625 pKa = 3.7TTTAPCGHH633 pKa = 7.46DD634 pKa = 3.06EE635 pKa = 4.35TTPSEE640 pKa = 4.37TDD642 pKa = 3.23TTPAALDD649 pKa = 3.51TTTPSGPVVTTPSGPEE665 pKa = 3.7TTTAPCGHH673 pKa = 7.46DD674 pKa = 3.06EE675 pKa = 4.35TTPSEE680 pKa = 4.37TDD682 pKa = 3.23TTPAALDD689 pKa = 3.51TTTPSGPVVTTPSGPEE705 pKa = 3.7TTTAPCGHH713 pKa = 7.46DD714 pKa = 3.06EE715 pKa = 4.35TTPSEE720 pKa = 4.37TDD722 pKa = 3.23TTPAALDD729 pKa = 3.51TTTPSGPVVTTPSGPEE745 pKa = 3.7TTTAPCGHH753 pKa = 7.46DD754 pKa = 3.06EE755 pKa = 4.35TTPSEE760 pKa = 4.37TDD762 pKa = 3.23TTPAALDD769 pKa = 3.51TTTPSGPVVTTPSGPEE785 pKa = 3.7TTTAPCGHH793 pKa = 7.46DD794 pKa = 3.06EE795 pKa = 4.35TTPSEE800 pKa = 4.37TDD802 pKa = 3.23TTPAALDD809 pKa = 3.51TTTPSGPVVTTPSGPEE825 pKa = 3.7TTTAPCGHH833 pKa = 7.46DD834 pKa = 3.06EE835 pKa = 4.35TTPSEE840 pKa = 4.37TDD842 pKa = 3.23TTPAALDD849 pKa = 3.51TTTPSGPVVTTPSGPEE865 pKa = 3.7TTTAPCGHH873 pKa = 7.46DD874 pKa = 3.06EE875 pKa = 4.35TTPSEE880 pKa = 4.37TDD882 pKa = 3.3TTPAAPDD889 pKa = 3.38TTTPIGPEE897 pKa = 3.31ITTPGGPEE905 pKa = 3.68VTTPGGPEE913 pKa = 3.29ITTPGGPEE921 pKa = 3.27ITTPGGPEE929 pKa = 3.27ITTPGGPEE937 pKa = 3.68VTTPGGPEE945 pKa = 3.68VTTPGGPEE953 pKa = 3.69VTTPSGPEE961 pKa = 3.24ITTPGGPEE969 pKa = 3.27ITTPGGPEE977 pKa = 3.68VTTPGGPEE985 pKa = 3.29ITTPGGPEE993 pKa = 3.46ITTSGGPEE1001 pKa = 3.43ITTPGGPEE1009 pKa = 3.58KK1010 pKa = 8.62TTPSGPEE1017 pKa = 3.68TTTAPCGHH1025 pKa = 7.46DD1026 pKa = 3.06EE1027 pKa = 4.35TTPSEE1032 pKa = 4.37TDD1034 pKa = 3.23TTPAALDD1041 pKa = 3.51TTTPSGPVVTTPSGPEE1057 pKa = 3.7TTTAPCGHH1065 pKa = 7.46DD1066 pKa = 3.06EE1067 pKa = 4.35TTPSEE1072 pKa = 4.37TDD1074 pKa = 3.23TTPAALDD1081 pKa = 3.47TTTPSGSVVTTPSGPEE1097 pKa = 3.7TTTAPCGHH1105 pKa = 7.46DD1106 pKa = 3.06EE1107 pKa = 4.35TTPSEE1112 pKa = 4.37TDD1114 pKa = 3.23TTPAALDD1121 pKa = 3.47TTTPSGSVVTTPSGPEE1137 pKa = 3.7TTTAPCGHH1145 pKa = 7.46DD1146 pKa = 3.06EE1147 pKa = 4.35TTPSEE1152 pKa = 4.37TDD1154 pKa = 3.23TTPAALDD1161 pKa = 3.49TTTPSGLVVTTPSGPEE1177 pKa = 3.7TTTAPCGHH1185 pKa = 7.46DD1186 pKa = 3.06EE1187 pKa = 4.35TTPSEE1192 pKa = 4.37TDD1194 pKa = 3.3TTPAAPDD1201 pKa = 3.41TTTPSGPVVTTPSGPEE1217 pKa = 3.7TTTAPCGHH1225 pKa = 7.46DD1226 pKa = 3.06EE1227 pKa = 4.35TTPSEE1232 pKa = 4.44TGTTPAAPDD1241 pKa = 3.01ITTPSGPEE1249 pKa = 3.69TTTAPCGHH1257 pKa = 7.46DD1258 pKa = 3.06EE1259 pKa = 4.35TTPSEE1264 pKa = 4.44TGTTPAAPDD1273 pKa = 3.45TTTPSGPVVTTPSGPEE1289 pKa = 3.7TTTAPCGHH1297 pKa = 7.46DD1298 pKa = 3.06EE1299 pKa = 4.35TTPSEE1304 pKa = 4.37TDD1306 pKa = 3.3TTPAAPDD1313 pKa = 3.41TTTPSGPVVTTPSGPEE1329 pKa = 3.7TTTAPCGHH1337 pKa = 7.46DD1338 pKa = 3.06EE1339 pKa = 4.35TTPSEE1344 pKa = 4.44TGTTPAAPDD1353 pKa = 3.45TTTPSGPEE1361 pKa = 3.77TTTAPCGHH1369 pKa = 7.46DD1370 pKa = 3.06EE1371 pKa = 4.35TTPSEE1376 pKa = 4.44TGTTPAAPDD1385 pKa = 3.45TTTPSGPVVTTPSGPEE1401 pKa = 3.7TTTAPCGHH1409 pKa = 7.46DD1410 pKa = 3.06EE1411 pKa = 4.35TTPSEE1416 pKa = 4.37TDD1418 pKa = 3.3TTPAAPDD1425 pKa = 3.41TTTPSGPVVTTPSGPEE1441 pKa = 3.7TTTAPCGHH1449 pKa = 7.46DD1450 pKa = 3.06EE1451 pKa = 4.35TTPSEE1456 pKa = 4.37TDD1458 pKa = 3.3TTPAAPDD1465 pKa = 3.33TATPSGPEE1473 pKa = 4.12TTTSPCGHH1481 pKa = 7.45DD1482 pKa = 3.12EE1483 pKa = 4.31TTPSEE1488 pKa = 4.37TDD1490 pKa = 3.23TTPAALDD1497 pKa = 3.49TTTPSGLVVTTPSGPEE1513 pKa = 3.7TTTAPCGHH1521 pKa = 7.49DD1522 pKa = 3.29EE1523 pKa = 4.29TTPSGPHH1530 pKa = 5.08VTTQSEE1536 pKa = 4.66PDD1538 pKa = 3.35TTTPRR1543 pKa = 11.84PATFPPITTPSSCQHH1558 pKa = 6.49HH1559 pKa = 6.47PTSEE1563 pKa = 4.07KK1564 pKa = 10.57PITSTSRR1571 pKa = 11.84PSTPPCGHH1579 pKa = 6.59SSTTKK1584 pKa = 8.97ATTIHH1589 pKa = 7.24RR1590 pKa = 11.84SGKK1593 pKa = 8.83PCKK1596 pKa = 10.08CSDD1599 pKa = 3.24GATTEE1604 pKa = 4.16VPTDD1608 pKa = 3.67VSDD1611 pKa = 3.94HH1612 pKa = 6.32LVDD1615 pKa = 3.84EE1616 pKa = 4.89TVHH1619 pKa = 6.38LLNTGVVAVEE1629 pKa = 4.35DD1630 pKa = 3.99LLEE1633 pKa = 4.23EE1634 pKa = 4.37LLASGQKK1641 pKa = 10.53DD1642 pKa = 3.0ILHH1645 pKa = 7.2KK1646 pKa = 10.24IRR1648 pKa = 11.84RR1649 pKa = 11.84TKK1651 pKa = 10.34RR1652 pKa = 11.84KK1653 pKa = 8.36NRR1655 pKa = 11.84RR1656 pKa = 11.84TRR1658 pKa = 11.84LHH1660 pKa = 6.51KK1661 pKa = 10.06PATVLPILII1670 pKa = 4.64
Molecular weight: 164.75 kDa
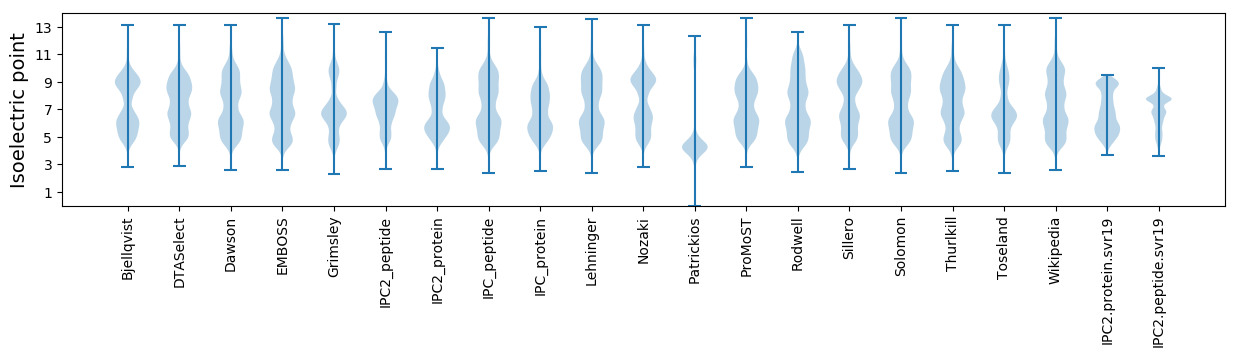
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2A4K6K5|A0A2A4K6K5_HELVI CRF domain-containing protein OS=Heliothis virescens OX=7102 GN=B5V51_13338 PE=4 SV=1
EEE2 pKa = 4.37AATASPYYY10 pKa = 10.67LAASLRR16 pKa = 11.84RR17 pKa = 11.84TASSYYY23 pKa = 11.15LAAPPRR29 pKa = 11.84RR30 pKa = 11.84TTAPHHH36 pKa = 6.78FAALPRR42 pKa = 11.84RR43 pKa = 11.84TTEEE47 pKa = 3.43HHH49 pKa = 6.65FATLPRR55 pKa = 11.84RR56 pKa = 11.84TNAPHHH62 pKa = 7.16FAALPRR68 pKa = 11.84RR69 pKa = 11.84TTAPHHH75 pKa = 6.89FAAPPRR81 pKa = 11.84RR82 pKa = 11.84TASPHHH88 pKa = 6.26LAAPPRR94 pKa = 11.84RR95 pKa = 11.84TASPHHH101 pKa = 6.19LAAPPHHH108 pKa = 5.94TASPHHH114 pKa = 5.78HH115 pKa = 6.47AAPLRR120 pKa = 11.84RR121 pKa = 11.84TASPHHH127 pKa = 6.73LTAPPRR133 pKa = 11.84RR134 pKa = 11.84TTAPHHH140 pKa = 6.89FAAPPRR146 pKa = 11.84RR147 pKa = 11.84TASPHHH153 pKa = 6.26LAAPPRR159 pKa = 11.84RR160 pKa = 11.84TASPHHH166 pKa = 6.19LAAPPHHH173 pKa = 5.94TASPHHH179 pKa = 5.78HH180 pKa = 6.47AAPLRR185 pKa = 11.84RR186 pKa = 11.84TASPHHH192 pKa = 6.73LTAPPRR198 pKa = 11.84RR199 pKa = 11.84TTAPHHH205 pKa = 6.89FAAPPRR211 pKa = 11.84RR212 pKa = 11.84TASPHHH218 pKa = 6.26LAAPPRR224 pKa = 11.84RR225 pKa = 11.84TASPHHH231 pKa = 6.19LAAPPHHH238 pKa = 5.94TASPHHH244 pKa = 5.78HH245 pKa = 6.47AAPLRR250 pKa = 11.84RR251 pKa = 11.84TASPHHH257 pKa = 6.73LTAPPRR263 pKa = 11.84RR264 pKa = 11.84TTAPHHH270 pKa = 6.89FAAPPRR276 pKa = 11.84RR277 pKa = 11.84TASPHHH283 pKa = 6.26LAAPPRR289 pKa = 11.84RR290 pKa = 11.84TASPHHH296 pKa = 6.19LAAPPHHH303 pKa = 5.94TASPHHH309 pKa = 5.78HH310 pKa = 6.47AAPLRR315 pKa = 11.84RR316 pKa = 11.84TASPHHH322 pKa = 6.73LTAPPRR328 pKa = 11.84RR329 pKa = 11.84TTAPHHH335 pKa = 6.89FAAPPRR341 pKa = 11.84RR342 pKa = 11.84TASPHHH348 pKa = 6.26LAAPPRR354 pKa = 11.84RR355 pKa = 11.84TASPHHH361 pKa = 6.19LAAPPHHH368 pKa = 5.94TASPHHH374 pKa = 5.78HH375 pKa = 6.47AAPLRR380 pKa = 11.84RR381 pKa = 11.84TASPHHH387 pKa = 6.73LTAPPRR393 pKa = 11.84RR394 pKa = 11.84TTAPHHH400 pKa = 6.89FAAPPRR406 pKa = 11.84RR407 pKa = 11.84TASPHHH413 pKa = 6.26LAAPPRR419 pKa = 11.84RR420 pKa = 11.84TASPHHH426 pKa = 6.19LAAPPHHH433 pKa = 5.94TASPHHH439 pKa = 5.78HH440 pKa = 6.47AAPLRR445 pKa = 11.84RR446 pKa = 11.84TASPHHH452 pKa = 6.73LTAPPRR458 pKa = 11.84RR459 pKa = 11.84TTAPHHH465 pKa = 6.89FAAPPRR471 pKa = 11.84RR472 pKa = 11.84TASPHHH478 pKa = 6.26LAAPPRR484 pKa = 11.84NPPSDDD490 pKa = 3.57PFAGRR495 pKa = 11.84IQRR498 pKa = 11.84MGEEE502 pKa = 3.95VVRR505 pKa = 11.84GRR507 pKa = 11.84
EEE2 pKa = 4.37AATASPYYY10 pKa = 10.67LAASLRR16 pKa = 11.84RR17 pKa = 11.84TASSYYY23 pKa = 11.15LAAPPRR29 pKa = 11.84RR30 pKa = 11.84TTAPHHH36 pKa = 6.78FAALPRR42 pKa = 11.84RR43 pKa = 11.84TTEEE47 pKa = 3.43HHH49 pKa = 6.65FATLPRR55 pKa = 11.84RR56 pKa = 11.84TNAPHHH62 pKa = 7.16FAALPRR68 pKa = 11.84RR69 pKa = 11.84TTAPHHH75 pKa = 6.89FAAPPRR81 pKa = 11.84RR82 pKa = 11.84TASPHHH88 pKa = 6.26LAAPPRR94 pKa = 11.84RR95 pKa = 11.84TASPHHH101 pKa = 6.19LAAPPHHH108 pKa = 5.94TASPHHH114 pKa = 5.78HH115 pKa = 6.47AAPLRR120 pKa = 11.84RR121 pKa = 11.84TASPHHH127 pKa = 6.73LTAPPRR133 pKa = 11.84RR134 pKa = 11.84TTAPHHH140 pKa = 6.89FAAPPRR146 pKa = 11.84RR147 pKa = 11.84TASPHHH153 pKa = 6.26LAAPPRR159 pKa = 11.84RR160 pKa = 11.84TASPHHH166 pKa = 6.19LAAPPHHH173 pKa = 5.94TASPHHH179 pKa = 5.78HH180 pKa = 6.47AAPLRR185 pKa = 11.84RR186 pKa = 11.84TASPHHH192 pKa = 6.73LTAPPRR198 pKa = 11.84RR199 pKa = 11.84TTAPHHH205 pKa = 6.89FAAPPRR211 pKa = 11.84RR212 pKa = 11.84TASPHHH218 pKa = 6.26LAAPPRR224 pKa = 11.84RR225 pKa = 11.84TASPHHH231 pKa = 6.19LAAPPHHH238 pKa = 5.94TASPHHH244 pKa = 5.78HH245 pKa = 6.47AAPLRR250 pKa = 11.84RR251 pKa = 11.84TASPHHH257 pKa = 6.73LTAPPRR263 pKa = 11.84RR264 pKa = 11.84TTAPHHH270 pKa = 6.89FAAPPRR276 pKa = 11.84RR277 pKa = 11.84TASPHHH283 pKa = 6.26LAAPPRR289 pKa = 11.84RR290 pKa = 11.84TASPHHH296 pKa = 6.19LAAPPHHH303 pKa = 5.94TASPHHH309 pKa = 5.78HH310 pKa = 6.47AAPLRR315 pKa = 11.84RR316 pKa = 11.84TASPHHH322 pKa = 6.73LTAPPRR328 pKa = 11.84RR329 pKa = 11.84TTAPHHH335 pKa = 6.89FAAPPRR341 pKa = 11.84RR342 pKa = 11.84TASPHHH348 pKa = 6.26LAAPPRR354 pKa = 11.84RR355 pKa = 11.84TASPHHH361 pKa = 6.19LAAPPHHH368 pKa = 5.94TASPHHH374 pKa = 5.78HH375 pKa = 6.47AAPLRR380 pKa = 11.84RR381 pKa = 11.84TASPHHH387 pKa = 6.73LTAPPRR393 pKa = 11.84RR394 pKa = 11.84TTAPHHH400 pKa = 6.89FAAPPRR406 pKa = 11.84RR407 pKa = 11.84TASPHHH413 pKa = 6.26LAAPPRR419 pKa = 11.84RR420 pKa = 11.84TASPHHH426 pKa = 6.19LAAPPHHH433 pKa = 5.94TASPHHH439 pKa = 5.78HH440 pKa = 6.47AAPLRR445 pKa = 11.84RR446 pKa = 11.84TASPHHH452 pKa = 6.73LTAPPRR458 pKa = 11.84RR459 pKa = 11.84TTAPHHH465 pKa = 6.89FAAPPRR471 pKa = 11.84RR472 pKa = 11.84TASPHHH478 pKa = 6.26LAAPPRR484 pKa = 11.84NPPSDDD490 pKa = 3.57PFAGRR495 pKa = 11.84IQRR498 pKa = 11.84MGEEE502 pKa = 3.95VVRR505 pKa = 11.84GRR507 pKa = 11.84
Molecular weight: 55.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
9040933 |
42 |
11599 |
530.1 |
59.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.453 ± 0.034 | 2.116 ± 0.041 |
5.622 ± 0.022 | 6.618 ± 0.034 |
3.312 ± 0.017 | 5.668 ± 0.034 |
2.56 ± 0.015 | 4.995 ± 0.02 |
6.168 ± 0.038 | 8.744 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.201 ± 0.012 | 4.534 ± 0.022 |
5.902 ± 0.047 | 4.002 ± 0.021 |
5.84 ± 0.027 | 7.629 ± 0.027 |
5.995 ± 0.038 | 6.435 ± 0.019 |
1.07 ± 0.009 | 3.122 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
