
Juglans regia (English walnut)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Fagales; Juglandaceae; Juglans
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
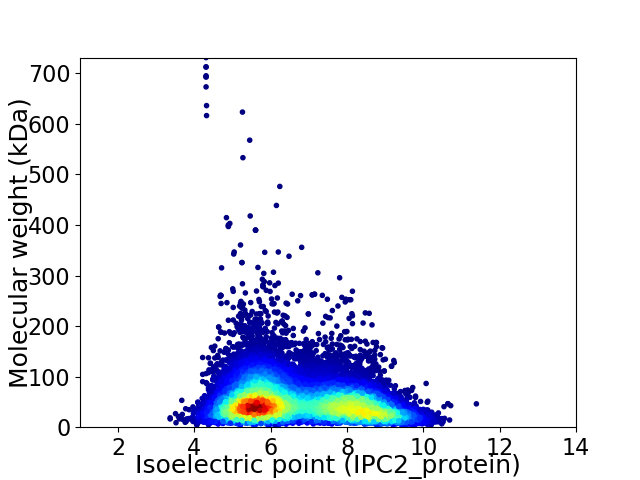
Virtual 2D-PAGE plot for 38418 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
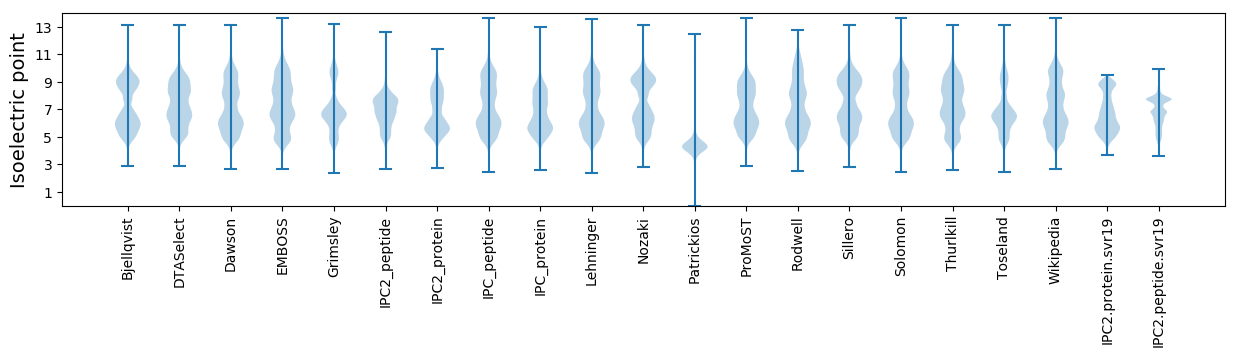
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2I4FJ54|A0A2I4FJ54_JUGRE FAM10 family protein At4g22670-like OS=Juglans regia OX=51240 GN=LOC108999267 PE=3 SV=1
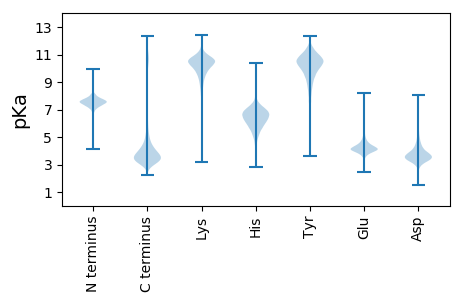
MM1 pKa = 7.53KK2 pKa = 10.18KK3 pKa = 10.33CEE5 pKa = 4.36LCDD8 pKa = 3.74SPANLYY14 pKa = 10.59CEE16 pKa = 4.52SDD18 pKa = 3.59QASLCWDD25 pKa = 3.52CDD27 pKa = 3.6ARR29 pKa = 11.84VHH31 pKa = 6.31GANFLVAKK39 pKa = 10.33HH40 pKa = 6.1SRR42 pKa = 11.84TLLCHH47 pKa = 6.07VCQSSTPWNGSGPKK61 pKa = 9.79LGPTISVCEE70 pKa = 3.68ICVNSNVKK78 pKa = 10.47NEE80 pKa = 4.27AGNEE84 pKa = 4.2GNDD87 pKa = 3.77HH88 pKa = 7.52DD89 pKa = 4.89NGADD93 pKa = 3.6GVGDD97 pKa = 4.23SDD99 pKa = 6.54DD100 pKa = 5.33DD101 pKa = 6.23DD102 pKa = 6.33SDD104 pKa = 4.59SDD106 pKa = 4.42DD107 pKa = 3.71NFEE110 pKa = 5.13EE111 pKa = 4.93EE112 pKa = 4.96EE113 pKa = 4.71DD114 pKa = 5.49DD115 pKa = 6.32DD116 pKa = 4.85GQGDD120 pKa = 3.68NDD122 pKa = 3.57EE123 pKa = 5.39DD124 pKa = 3.99EE125 pKa = 5.19EE126 pKa = 4.47EE127 pKa = 4.33NQVVPWSSTPPPPTSSSSWRR147 pKa = 11.84GIHH150 pKa = 6.58
MM1 pKa = 7.53KK2 pKa = 10.18KK3 pKa = 10.33CEE5 pKa = 4.36LCDD8 pKa = 3.74SPANLYY14 pKa = 10.59CEE16 pKa = 4.52SDD18 pKa = 3.59QASLCWDD25 pKa = 3.52CDD27 pKa = 3.6ARR29 pKa = 11.84VHH31 pKa = 6.31GANFLVAKK39 pKa = 10.33HH40 pKa = 6.1SRR42 pKa = 11.84TLLCHH47 pKa = 6.07VCQSSTPWNGSGPKK61 pKa = 9.79LGPTISVCEE70 pKa = 3.68ICVNSNVKK78 pKa = 10.47NEE80 pKa = 4.27AGNEE84 pKa = 4.2GNDD87 pKa = 3.77HH88 pKa = 7.52DD89 pKa = 4.89NGADD93 pKa = 3.6GVGDD97 pKa = 4.23SDD99 pKa = 6.54DD100 pKa = 5.33DD101 pKa = 6.23DD102 pKa = 6.33SDD104 pKa = 4.59SDD106 pKa = 4.42DD107 pKa = 3.71NFEE110 pKa = 5.13EE111 pKa = 4.93EE112 pKa = 4.96EE113 pKa = 4.71DD114 pKa = 5.49DD115 pKa = 6.32DD116 pKa = 4.85GQGDD120 pKa = 3.68NDD122 pKa = 3.57EE123 pKa = 5.39DD124 pKa = 3.99EE125 pKa = 5.19EE126 pKa = 4.47EE127 pKa = 4.33NQVVPWSSTPPPPTSSSSWRR147 pKa = 11.84GIHH150 pKa = 6.58
Molecular weight: 16.21 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6P9DV69|A0A6P9DV69_JUGRE uncharacterized protein LOC109011743 OS=Juglans regia OX=51240 GN=LOC109011743 PE=4 SV=1
MM1 pKa = 7.48AHH3 pKa = 7.41LLLEE7 pKa = 4.52SLQKK11 pKa = 10.48GAPISPSVPNPTIPPSLSTISQRR34 pKa = 11.84TFTSHH39 pKa = 6.47PRR41 pKa = 11.84AHH43 pKa = 7.11LLGTVLQKK51 pKa = 10.94GHH53 pKa = 7.07IPPSAPNPPIPPRR66 pKa = 11.84LSTISQRR73 pKa = 11.84AFASHH78 pKa = 6.85PRR80 pKa = 11.84AHH82 pKa = 6.86LLWTILQRR90 pKa = 11.84GHH92 pKa = 6.96IPPPAPNPTIPPRR105 pKa = 11.84LSTINQKK112 pKa = 10.77AFGSHH117 pKa = 6.0PRR119 pKa = 11.84AHH121 pKa = 7.12LLGTVLQKK129 pKa = 10.91GHH131 pKa = 7.05IPPPGPNSKK140 pKa = 9.78IPRR143 pKa = 11.84HH144 pKa = 6.05LSTISQRR151 pKa = 11.84AFASHH156 pKa = 6.86PRR158 pKa = 11.84AHH160 pKa = 7.2LLGTVLQRR168 pKa = 11.84GHH170 pKa = 6.97IPPSVPNPPILPRR183 pKa = 11.84LSTISQRR190 pKa = 11.84AFASHH195 pKa = 6.85PRR197 pKa = 11.84AHH199 pKa = 6.86LLWTILQRR207 pKa = 11.84GPIPPPAPNPTIPPRR222 pKa = 11.84LSTINQKK229 pKa = 10.77AFGSHH234 pKa = 6.0PRR236 pKa = 11.84AHH238 pKa = 7.12LLGTVLQKK246 pKa = 10.91GHH248 pKa = 7.05IPPPGPNSKK257 pKa = 9.78IPRR260 pKa = 11.84HH261 pKa = 6.05LSTISQRR268 pKa = 11.84AFASHH273 pKa = 6.86PRR275 pKa = 11.84AHH277 pKa = 7.2LLGTVLQRR285 pKa = 11.84GHH287 pKa = 6.97IPPSVPNPTIPPRR300 pKa = 11.84FSTISQRR307 pKa = 11.84AFASHH312 pKa = 6.85PRR314 pKa = 11.84AHH316 pKa = 6.86LLWTVLQRR324 pKa = 11.84GHH326 pKa = 6.95IPPPAPNPTIPPRR339 pKa = 11.84LSTINQKK346 pKa = 10.77AFGSHH351 pKa = 6.0PRR353 pKa = 11.84AHH355 pKa = 7.09LLGTILQKK363 pKa = 10.59SHH365 pKa = 7.03IPPSAPNPTIPPLLSTINQKK385 pKa = 10.59AFASHH390 pKa = 6.77PRR392 pKa = 11.84AHH394 pKa = 7.2LLGTVLQKK402 pKa = 10.92GHH404 pKa = 7.03IPPSGPNSKK413 pKa = 9.73IPRR416 pKa = 11.84HH417 pKa = 5.69ISTLISQNNNN427 pKa = 2.76
MM1 pKa = 7.48AHH3 pKa = 7.41LLLEE7 pKa = 4.52SLQKK11 pKa = 10.48GAPISPSVPNPTIPPSLSTISQRR34 pKa = 11.84TFTSHH39 pKa = 6.47PRR41 pKa = 11.84AHH43 pKa = 7.11LLGTVLQKK51 pKa = 10.94GHH53 pKa = 7.07IPPSAPNPPIPPRR66 pKa = 11.84LSTISQRR73 pKa = 11.84AFASHH78 pKa = 6.85PRR80 pKa = 11.84AHH82 pKa = 6.86LLWTILQRR90 pKa = 11.84GHH92 pKa = 6.96IPPPAPNPTIPPRR105 pKa = 11.84LSTINQKK112 pKa = 10.77AFGSHH117 pKa = 6.0PRR119 pKa = 11.84AHH121 pKa = 7.12LLGTVLQKK129 pKa = 10.91GHH131 pKa = 7.05IPPPGPNSKK140 pKa = 9.78IPRR143 pKa = 11.84HH144 pKa = 6.05LSTISQRR151 pKa = 11.84AFASHH156 pKa = 6.86PRR158 pKa = 11.84AHH160 pKa = 7.2LLGTVLQRR168 pKa = 11.84GHH170 pKa = 6.97IPPSVPNPPILPRR183 pKa = 11.84LSTISQRR190 pKa = 11.84AFASHH195 pKa = 6.85PRR197 pKa = 11.84AHH199 pKa = 6.86LLWTILQRR207 pKa = 11.84GPIPPPAPNPTIPPRR222 pKa = 11.84LSTINQKK229 pKa = 10.77AFGSHH234 pKa = 6.0PRR236 pKa = 11.84AHH238 pKa = 7.12LLGTVLQKK246 pKa = 10.91GHH248 pKa = 7.05IPPPGPNSKK257 pKa = 9.78IPRR260 pKa = 11.84HH261 pKa = 6.05LSTISQRR268 pKa = 11.84AFASHH273 pKa = 6.86PRR275 pKa = 11.84AHH277 pKa = 7.2LLGTVLQRR285 pKa = 11.84GHH287 pKa = 6.97IPPSVPNPTIPPRR300 pKa = 11.84FSTISQRR307 pKa = 11.84AFASHH312 pKa = 6.85PRR314 pKa = 11.84AHH316 pKa = 6.86LLWTVLQRR324 pKa = 11.84GHH326 pKa = 6.95IPPPAPNPTIPPRR339 pKa = 11.84LSTINQKK346 pKa = 10.77AFGSHH351 pKa = 6.0PRR353 pKa = 11.84AHH355 pKa = 7.09LLGTILQKK363 pKa = 10.59SHH365 pKa = 7.03IPPSAPNPTIPPLLSTINQKK385 pKa = 10.59AFASHH390 pKa = 6.77PRR392 pKa = 11.84AHH394 pKa = 7.2LLGTVLQKK402 pKa = 10.92GHH404 pKa = 7.03IPPSGPNSKK413 pKa = 9.73IPRR416 pKa = 11.84HH417 pKa = 5.69ISTLISQNNNN427 pKa = 2.76
Molecular weight: 46.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
18253930 |
17 |
6605 |
475.1 |
53.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.634 ± 0.011 | 1.878 ± 0.006 |
5.328 ± 0.008 | 6.605 ± 0.022 |
4.17 ± 0.009 | 6.539 ± 0.01 |
2.439 ± 0.005 | 5.326 ± 0.01 |
6.025 ± 0.012 | 9.783 ± 0.017 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.405 ± 0.005 | 4.449 ± 0.008 |
4.901 ± 0.011 | 3.745 ± 0.008 |
5.39 ± 0.011 | 9.036 ± 0.014 |
4.848 ± 0.008 | 6.423 ± 0.008 |
1.302 ± 0.005 | 2.773 ± 0.007 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
