
bacterium HR21
Taxonomy: cellular organisms; Bacteria; unclassified Bacteria
Average proteome isoelectric point is 7.03
Get precalculated fractions of proteins
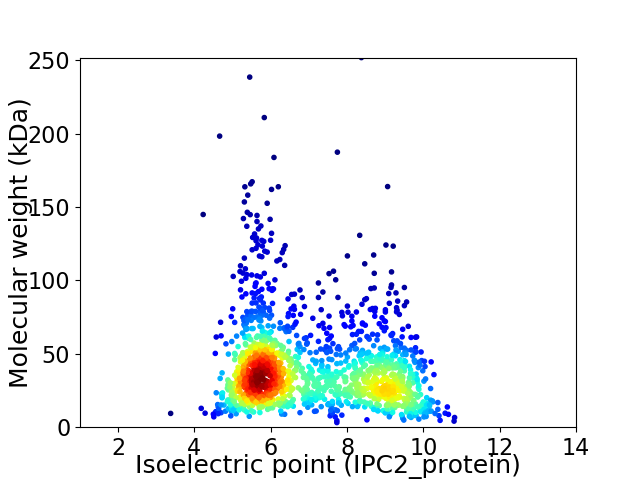
Virtual 2D-PAGE plot for 1558 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H5Y2I5|A0A2H5Y2I5_9BACT Thiol-disulfide oxidoreductase ResA OS=bacterium HR21 OX=2035416 GN=resA_5 PE=4 SV=1
MM1 pKa = 7.33FWTPEE6 pKa = 3.57LAAWLEE12 pKa = 4.16DD13 pKa = 4.07APWPATKK20 pKa = 10.09EE21 pKa = 4.02VLLDD25 pKa = 3.38YY26 pKa = 10.94AEE28 pKa = 4.71RR29 pKa = 11.84SGAPLQVIEE38 pKa = 4.59NLKK41 pKa = 10.11EE42 pKa = 4.05LPDD45 pKa = 5.25DD46 pKa = 3.73DD47 pKa = 4.5TEE49 pKa = 4.68YY50 pKa = 11.29EE51 pKa = 4.49GIEE54 pKa = 4.58DD55 pKa = 4.63IWPEE59 pKa = 3.57WADD62 pKa = 3.65FQLPDD67 pKa = 3.92YY68 pKa = 10.83LDD70 pKa = 3.86EE71 pKa = 6.18DD72 pKa = 3.92EE73 pKa = 5.49EE74 pKa = 4.33EE75 pKa = 4.29QEE77 pKa = 4.44EE78 pKa = 4.78YY79 pKa = 11.37
MM1 pKa = 7.33FWTPEE6 pKa = 3.57LAAWLEE12 pKa = 4.16DD13 pKa = 4.07APWPATKK20 pKa = 10.09EE21 pKa = 4.02VLLDD25 pKa = 3.38YY26 pKa = 10.94AEE28 pKa = 4.71RR29 pKa = 11.84SGAPLQVIEE38 pKa = 4.59NLKK41 pKa = 10.11EE42 pKa = 4.05LPDD45 pKa = 5.25DD46 pKa = 3.73DD47 pKa = 4.5TEE49 pKa = 4.68YY50 pKa = 11.29EE51 pKa = 4.49GIEE54 pKa = 4.58DD55 pKa = 4.63IWPEE59 pKa = 3.57WADD62 pKa = 3.65FQLPDD67 pKa = 3.92YY68 pKa = 10.83LDD70 pKa = 3.86EE71 pKa = 6.18DD72 pKa = 3.92EE73 pKa = 5.49EE74 pKa = 4.33EE75 pKa = 4.29QEE77 pKa = 4.44EE78 pKa = 4.78YY79 pKa = 11.37
Molecular weight: 9.39 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H5Y266|A0A2H5Y266_9BACT Uncharacterized protein OS=bacterium HR21 OX=2035416 GN=HRbin21_01341 PE=4 SV=1
MM1 pKa = 7.37EE2 pKa = 5.7RR3 pKa = 11.84SSNAISRR10 pKa = 11.84RR11 pKa = 11.84PMPGLCRR18 pKa = 11.84HH19 pKa = 5.45SCYY22 pKa = 10.43LGLLIFLAGIVPLWGWCSRR41 pKa = 11.84EE42 pKa = 4.11DD43 pKa = 3.36STAALSALRR52 pKa = 11.84ARR54 pKa = 11.84VQALVEE60 pKa = 3.93QLQQRR65 pKa = 11.84ARR67 pKa = 11.84VGVLVSDD74 pKa = 3.92VAGRR78 pKa = 11.84RR79 pKa = 11.84YY80 pKa = 9.75FEE82 pKa = 5.09HH83 pKa = 6.97GAEE86 pKa = 4.0QLLVPASVTKK96 pKa = 10.21LFWSVAALAVLGDD109 pKa = 3.79SAVIRR114 pKa = 11.84TALYY118 pKa = 10.89ADD120 pKa = 4.33ALPRR124 pKa = 11.84ADD126 pKa = 3.7GTLSGNLYY134 pKa = 10.68LVGGGDD140 pKa = 3.81ALLTVQEE147 pKa = 4.52LEE149 pKa = 4.03EE150 pKa = 4.25LAAQLVRR157 pKa = 11.84RR158 pKa = 11.84GVRR161 pKa = 11.84RR162 pKa = 11.84ITGAVLGDD170 pKa = 3.33GTLFDD175 pKa = 4.13GQRR178 pKa = 11.84TRR180 pKa = 11.84LQYY183 pKa = 11.04SGDD186 pKa = 3.79SDD188 pKa = 3.99PVEE191 pKa = 4.31PLPPITALGFNRR203 pKa = 11.84NEE205 pKa = 3.62VRR207 pKa = 11.84VLVSATGRR215 pKa = 11.84RR216 pKa = 11.84VTAQTIPVSPGFVVDD231 pKa = 3.58VSGVRR236 pKa = 11.84IRR238 pKa = 11.84KK239 pKa = 6.88GARR242 pKa = 11.84RR243 pKa = 11.84GRR245 pKa = 11.84QRR247 pKa = 11.84VVLSARR253 pKa = 11.84SVMRR257 pKa = 11.84RR258 pKa = 11.84GVQYY262 pKa = 10.37IVVSGWIAARR272 pKa = 11.84GTWSLGVPMEE282 pKa = 4.33KK283 pKa = 10.28PEE285 pKa = 4.0VAAAATLVQRR295 pKa = 11.84LRR297 pKa = 11.84AAGIRR302 pKa = 11.84VEE304 pKa = 4.35GGFGEE309 pKa = 5.22GKK311 pKa = 9.98CPPGATLLGLWEE323 pKa = 4.55RR324 pKa = 11.84PLAEE328 pKa = 5.01LLAAVNKK335 pKa = 10.48EE336 pKa = 3.5SDD338 pKa = 3.57NFVAEE343 pKa = 4.52HH344 pKa = 6.59LFKK347 pKa = 11.32LLGARR352 pKa = 11.84AAGSGTHH359 pKa = 6.58AEE361 pKa = 4.06RR362 pKa = 11.84ARR364 pKa = 11.84RR365 pKa = 11.84LLQALLSRR373 pKa = 11.84WGIPCQGCALWDD385 pKa = 3.76GSGLSRR391 pKa = 11.84RR392 pKa = 11.84NRR394 pKa = 11.84VSAADD399 pKa = 3.49VVGMLRR405 pKa = 11.84AVFQLPIADD414 pKa = 3.92ALCRR418 pKa = 11.84SLSIAGVDD426 pKa = 3.54GTLKK430 pKa = 10.28QRR432 pKa = 11.84LQQTQAQARR441 pKa = 11.84VWAKK445 pKa = 10.11TGTLRR450 pKa = 11.84NASALAGYY458 pKa = 10.25VLTADD463 pKa = 4.04GDD465 pKa = 4.23PLVFAILSFGVVSRR479 pKa = 11.84AKK481 pKa = 10.46ALEE484 pKa = 4.4DD485 pKa = 3.62SLVATLARR493 pKa = 11.84FSFCGAGTQSRR504 pKa = 3.91
MM1 pKa = 7.37EE2 pKa = 5.7RR3 pKa = 11.84SSNAISRR10 pKa = 11.84RR11 pKa = 11.84PMPGLCRR18 pKa = 11.84HH19 pKa = 5.45SCYY22 pKa = 10.43LGLLIFLAGIVPLWGWCSRR41 pKa = 11.84EE42 pKa = 4.11DD43 pKa = 3.36STAALSALRR52 pKa = 11.84ARR54 pKa = 11.84VQALVEE60 pKa = 3.93QLQQRR65 pKa = 11.84ARR67 pKa = 11.84VGVLVSDD74 pKa = 3.92VAGRR78 pKa = 11.84RR79 pKa = 11.84YY80 pKa = 9.75FEE82 pKa = 5.09HH83 pKa = 6.97GAEE86 pKa = 4.0QLLVPASVTKK96 pKa = 10.21LFWSVAALAVLGDD109 pKa = 3.79SAVIRR114 pKa = 11.84TALYY118 pKa = 10.89ADD120 pKa = 4.33ALPRR124 pKa = 11.84ADD126 pKa = 3.7GTLSGNLYY134 pKa = 10.68LVGGGDD140 pKa = 3.81ALLTVQEE147 pKa = 4.52LEE149 pKa = 4.03EE150 pKa = 4.25LAAQLVRR157 pKa = 11.84RR158 pKa = 11.84GVRR161 pKa = 11.84RR162 pKa = 11.84ITGAVLGDD170 pKa = 3.33GTLFDD175 pKa = 4.13GQRR178 pKa = 11.84TRR180 pKa = 11.84LQYY183 pKa = 11.04SGDD186 pKa = 3.79SDD188 pKa = 3.99PVEE191 pKa = 4.31PLPPITALGFNRR203 pKa = 11.84NEE205 pKa = 3.62VRR207 pKa = 11.84VLVSATGRR215 pKa = 11.84RR216 pKa = 11.84VTAQTIPVSPGFVVDD231 pKa = 3.58VSGVRR236 pKa = 11.84IRR238 pKa = 11.84KK239 pKa = 6.88GARR242 pKa = 11.84RR243 pKa = 11.84GRR245 pKa = 11.84QRR247 pKa = 11.84VVLSARR253 pKa = 11.84SVMRR257 pKa = 11.84RR258 pKa = 11.84GVQYY262 pKa = 10.37IVVSGWIAARR272 pKa = 11.84GTWSLGVPMEE282 pKa = 4.33KK283 pKa = 10.28PEE285 pKa = 4.0VAAAATLVQRR295 pKa = 11.84LRR297 pKa = 11.84AAGIRR302 pKa = 11.84VEE304 pKa = 4.35GGFGEE309 pKa = 5.22GKK311 pKa = 9.98CPPGATLLGLWEE323 pKa = 4.55RR324 pKa = 11.84PLAEE328 pKa = 5.01LLAAVNKK335 pKa = 10.48EE336 pKa = 3.5SDD338 pKa = 3.57NFVAEE343 pKa = 4.52HH344 pKa = 6.59LFKK347 pKa = 11.32LLGARR352 pKa = 11.84AAGSGTHH359 pKa = 6.58AEE361 pKa = 4.06RR362 pKa = 11.84ARR364 pKa = 11.84RR365 pKa = 11.84LLQALLSRR373 pKa = 11.84WGIPCQGCALWDD385 pKa = 3.76GSGLSRR391 pKa = 11.84RR392 pKa = 11.84NRR394 pKa = 11.84VSAADD399 pKa = 3.49VVGMLRR405 pKa = 11.84AVFQLPIADD414 pKa = 3.92ALCRR418 pKa = 11.84SLSIAGVDD426 pKa = 3.54GTLKK430 pKa = 10.28QRR432 pKa = 11.84LQQTQAQARR441 pKa = 11.84VWAKK445 pKa = 10.11TGTLRR450 pKa = 11.84NASALAGYY458 pKa = 10.25VLTADD463 pKa = 4.04GDD465 pKa = 4.23PLVFAILSFGVVSRR479 pKa = 11.84AKK481 pKa = 10.46ALEE484 pKa = 4.4DD485 pKa = 3.62SLVATLARR493 pKa = 11.84FSFCGAGTQSRR504 pKa = 3.91
Molecular weight: 53.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
581441 |
29 |
2214 |
373.2 |
41.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.005 ± 0.069 | 1.016 ± 0.019 |
3.84 ± 0.039 | 6.909 ± 0.075 |
3.747 ± 0.037 | 7.686 ± 0.069 |
2.211 ± 0.03 | 4.286 ± 0.043 |
1.818 ± 0.037 | 12.464 ± 0.083 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.541 ± 0.023 | 2.012 ± 0.036 |
6.08 ± 0.05 | 4.396 ± 0.047 |
8.913 ± 0.062 | 5.115 ± 0.046 |
5.129 ± 0.052 | 8.012 ± 0.061 |
1.89 ± 0.03 | 2.929 ± 0.038 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
