
Jannaschia donghaensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Jannaschia
Average proteome isoelectric point is 6.09
Get precalculated fractions of proteins
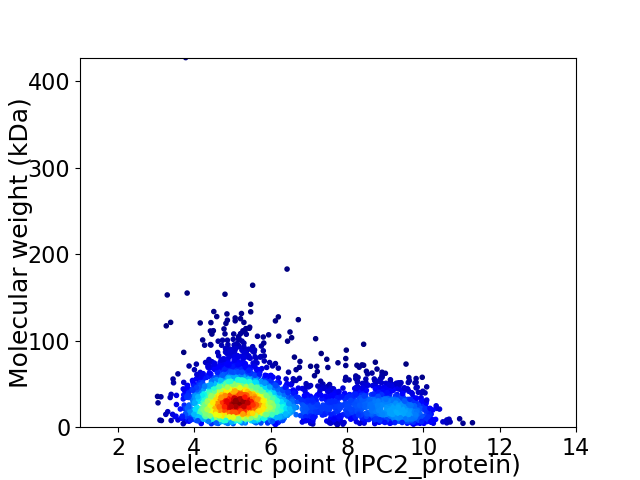
Virtual 2D-PAGE plot for 3480 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0M6YE48|A0A0M6YE48_9RHOB Succinate-semialdehyde dehydrogenase [NADP(+)] 1 OS=Jannaschia donghaensis OX=420998 GN=gabD1 PE=4 SV=1
MM1 pKa = 7.13TRR3 pKa = 11.84SKK5 pKa = 10.91FLGSTFCLASLLAAPTTAQEE25 pKa = 4.3RR26 pKa = 11.84PLTVAMHH33 pKa = 5.54YY34 pKa = 7.74TQEE37 pKa = 3.85QAAPLIACLDD47 pKa = 3.77AYY49 pKa = 9.95GTDD52 pKa = 3.93GPGAEE57 pKa = 4.37YY58 pKa = 10.44QQISYY63 pKa = 11.06GDD65 pKa = 3.65YY66 pKa = 10.52LQTVLTGRR74 pKa = 11.84LAGQAPDD81 pKa = 3.21IYY83 pKa = 10.92NVYY86 pKa = 10.21SIWAAQMVDD95 pKa = 3.24NGVLATPPAAVTDD108 pKa = 4.44FVTAGYY114 pKa = 9.97SAGTVDD120 pKa = 4.69AATIDD125 pKa = 3.68GTLWGVPTEE134 pKa = 4.05VSVYY138 pKa = 9.89MLVSNMALLRR148 pKa = 11.84EE149 pKa = 4.57AGFDD153 pKa = 3.67TPPTTWAEE161 pKa = 4.06MRR163 pKa = 11.84EE164 pKa = 4.03MAQAITTRR172 pKa = 11.84NDD174 pKa = 2.67QDD176 pKa = 3.93RR177 pKa = 11.84IEE179 pKa = 4.22TAGFAFAQSSSGAGLVHH196 pKa = 7.03PFYY199 pKa = 11.23ALTYY203 pKa = 10.15SEE205 pKa = 5.37GGDD208 pKa = 3.0IYY210 pKa = 11.4ADD212 pKa = 3.69DD213 pKa = 4.35RR214 pKa = 11.84SEE216 pKa = 5.37AMLEE220 pKa = 3.92SDD222 pKa = 4.87AATATAGLMAGMVQDD237 pKa = 5.55GITDD241 pKa = 4.32LSVDD245 pKa = 4.04AYY247 pKa = 10.57DD248 pKa = 5.01FPAGGIGMMIMANWYY263 pKa = 9.02EE264 pKa = 4.05SAIRR268 pKa = 11.84EE269 pKa = 4.37GFGDD273 pKa = 4.53RR274 pKa = 11.84FDD276 pKa = 4.47EE277 pKa = 4.54DD278 pKa = 4.12VAVSAIPMGEE288 pKa = 4.04DD289 pKa = 2.85WKK291 pKa = 9.71TMQYY295 pKa = 11.21AFFMGVDD302 pKa = 3.76SASDD306 pKa = 3.25RR307 pKa = 11.84SDD309 pKa = 3.51AAWDD313 pKa = 3.67LVRR316 pKa = 11.84HH317 pKa = 5.6LNEE320 pKa = 4.6PRR322 pKa = 11.84DD323 pKa = 3.86GAPSCMAEE331 pKa = 4.04MLDD334 pKa = 4.15GLGALTASSADD345 pKa = 3.41TAALPEE351 pKa = 4.67ADD353 pKa = 4.74AFTAPFVAALADD365 pKa = 3.9DD366 pKa = 4.32RR367 pKa = 11.84AISQPNVMQASEE379 pKa = 4.63LEE381 pKa = 4.08GLIARR386 pKa = 11.84TLEE389 pKa = 3.93EE390 pKa = 4.64VIVGDD395 pKa = 4.37AEE397 pKa = 4.32AGPAMEE403 pKa = 5.11EE404 pKa = 3.84LDD406 pKa = 4.28GDD408 pKa = 4.09VEE410 pKa = 5.22DD411 pKa = 4.87ILSEE415 pKa = 4.68FYY417 pKa = 11.22
MM1 pKa = 7.13TRR3 pKa = 11.84SKK5 pKa = 10.91FLGSTFCLASLLAAPTTAQEE25 pKa = 4.3RR26 pKa = 11.84PLTVAMHH33 pKa = 5.54YY34 pKa = 7.74TQEE37 pKa = 3.85QAAPLIACLDD47 pKa = 3.77AYY49 pKa = 9.95GTDD52 pKa = 3.93GPGAEE57 pKa = 4.37YY58 pKa = 10.44QQISYY63 pKa = 11.06GDD65 pKa = 3.65YY66 pKa = 10.52LQTVLTGRR74 pKa = 11.84LAGQAPDD81 pKa = 3.21IYY83 pKa = 10.92NVYY86 pKa = 10.21SIWAAQMVDD95 pKa = 3.24NGVLATPPAAVTDD108 pKa = 4.44FVTAGYY114 pKa = 9.97SAGTVDD120 pKa = 4.69AATIDD125 pKa = 3.68GTLWGVPTEE134 pKa = 4.05VSVYY138 pKa = 9.89MLVSNMALLRR148 pKa = 11.84EE149 pKa = 4.57AGFDD153 pKa = 3.67TPPTTWAEE161 pKa = 4.06MRR163 pKa = 11.84EE164 pKa = 4.03MAQAITTRR172 pKa = 11.84NDD174 pKa = 2.67QDD176 pKa = 3.93RR177 pKa = 11.84IEE179 pKa = 4.22TAGFAFAQSSSGAGLVHH196 pKa = 7.03PFYY199 pKa = 11.23ALTYY203 pKa = 10.15SEE205 pKa = 5.37GGDD208 pKa = 3.0IYY210 pKa = 11.4ADD212 pKa = 3.69DD213 pKa = 4.35RR214 pKa = 11.84SEE216 pKa = 5.37AMLEE220 pKa = 3.92SDD222 pKa = 4.87AATATAGLMAGMVQDD237 pKa = 5.55GITDD241 pKa = 4.32LSVDD245 pKa = 4.04AYY247 pKa = 10.57DD248 pKa = 5.01FPAGGIGMMIMANWYY263 pKa = 9.02EE264 pKa = 4.05SAIRR268 pKa = 11.84EE269 pKa = 4.37GFGDD273 pKa = 4.53RR274 pKa = 11.84FDD276 pKa = 4.47EE277 pKa = 4.54DD278 pKa = 4.12VAVSAIPMGEE288 pKa = 4.04DD289 pKa = 2.85WKK291 pKa = 9.71TMQYY295 pKa = 11.21AFFMGVDD302 pKa = 3.76SASDD306 pKa = 3.25RR307 pKa = 11.84SDD309 pKa = 3.51AAWDD313 pKa = 3.67LVRR316 pKa = 11.84HH317 pKa = 5.6LNEE320 pKa = 4.6PRR322 pKa = 11.84DD323 pKa = 3.86GAPSCMAEE331 pKa = 4.04MLDD334 pKa = 4.15GLGALTASSADD345 pKa = 3.41TAALPEE351 pKa = 4.67ADD353 pKa = 4.74AFTAPFVAALADD365 pKa = 3.9DD366 pKa = 4.32RR367 pKa = 11.84AISQPNVMQASEE379 pKa = 4.63LEE381 pKa = 4.08GLIARR386 pKa = 11.84TLEE389 pKa = 3.93EE390 pKa = 4.64VIVGDD395 pKa = 4.37AEE397 pKa = 4.32AGPAMEE403 pKa = 5.11EE404 pKa = 3.84LDD406 pKa = 4.28GDD408 pKa = 4.09VEE410 pKa = 5.22DD411 pKa = 4.87ILSEE415 pKa = 4.68FYY417 pKa = 11.22
Molecular weight: 44.29 kDa
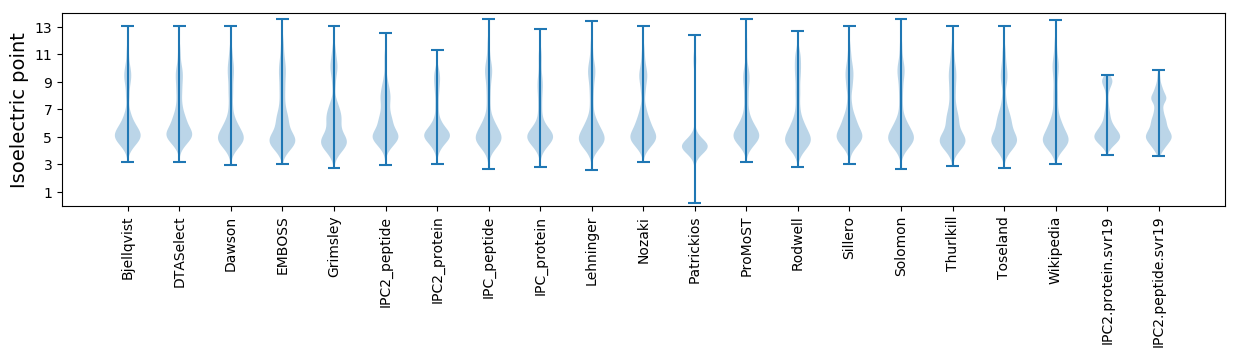
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0M6YI42|A0A0M6YI42_9RHOB YGGT family protein OS=Jannaschia donghaensis OX=420998 GN=JDO7802_00945 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.08AGRR28 pKa = 11.84RR29 pKa = 11.84ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.08AGRR28 pKa = 11.84RR29 pKa = 11.84ILNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1044310 |
30 |
4206 |
300.1 |
32.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.849 ± 0.059 | 0.86 ± 0.013 |
6.893 ± 0.045 | 5.213 ± 0.045 |
3.581 ± 0.025 | 8.957 ± 0.046 |
2.014 ± 0.022 | 5.02 ± 0.029 |
2.648 ± 0.038 | 9.965 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.795 ± 0.021 | 2.307 ± 0.023 |
5.247 ± 0.031 | 2.917 ± 0.023 |
7.034 ± 0.051 | 4.707 ± 0.029 |
6.025 ± 0.046 | 7.533 ± 0.038 |
1.444 ± 0.02 | 1.992 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
