
Roseovarius aestuarii
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Roseovarius
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
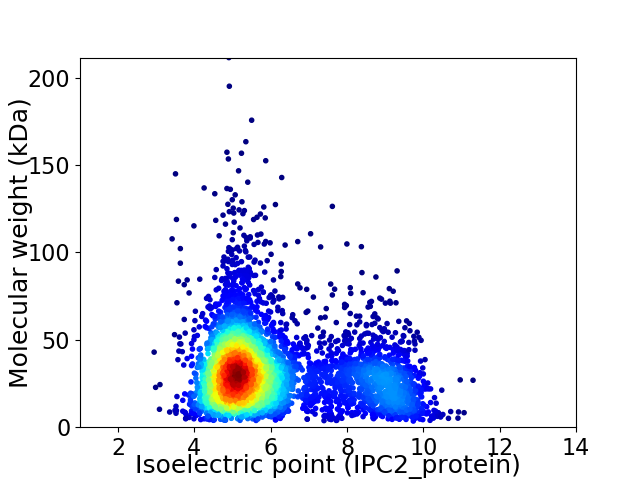
Virtual 2D-PAGE plot for 4594 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X7BX86|A0A1X7BX86_9RHOB Ferric uptake regulation protein OS=Roseovarius aestuarii OX=475083 GN=zur_2 PE=3 SV=1
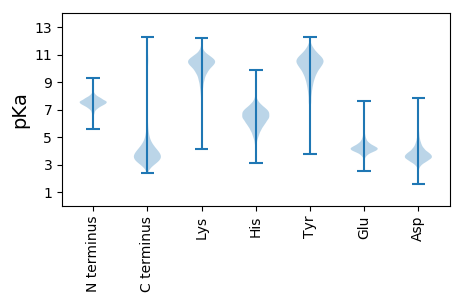
MM1 pKa = 7.89RR2 pKa = 11.84ISHH5 pKa = 7.57FYY7 pKa = 10.06TDD9 pKa = 3.56ADD11 pKa = 3.77EE12 pKa = 5.84KK13 pKa = 10.4IFGYY17 pKa = 10.09ISYY20 pKa = 10.05GWTFQNGQWVLGLTFRR36 pKa = 11.84DD37 pKa = 4.08DD38 pKa = 4.18EE39 pKa = 4.25IHH41 pKa = 7.3AGGGDD46 pKa = 3.74DD47 pKa = 4.31TIKK50 pKa = 10.99SSGGDD55 pKa = 3.44DD56 pKa = 3.08IVYY59 pKa = 9.26TGAGADD65 pKa = 3.67FVDD68 pKa = 4.16AGEE71 pKa = 4.72GNDD74 pKa = 4.45TIYY77 pKa = 11.45GDD79 pKa = 3.75AGEE82 pKa = 4.05DD83 pKa = 3.48TIYY86 pKa = 11.19GGDD89 pKa = 3.82GNDD92 pKa = 4.73FIYY95 pKa = 10.51GSPQGGLFYY104 pKa = 11.05GGDD107 pKa = 3.79GNDD110 pKa = 4.03FISGEE115 pKa = 4.24GAAHH119 pKa = 6.0VSIFGDD125 pKa = 3.35AGNDD129 pKa = 3.64TLVGGSGSDD138 pKa = 3.42TVIGGIGNDD147 pKa = 3.25TLYY150 pKa = 9.49ITEE153 pKa = 4.86GQDD156 pKa = 3.16NMHH159 pKa = 6.68GGEE162 pKa = 4.26GDD164 pKa = 3.82DD165 pKa = 5.58VFILDD170 pKa = 4.64QSVADD175 pKa = 3.96LGFRR179 pKa = 11.84TSVFDD184 pKa = 3.82YY185 pKa = 10.84FDD187 pKa = 3.73GSEE190 pKa = 4.71GYY192 pKa = 7.8DD193 pKa = 3.51TLDD196 pKa = 3.04MSAASGRR203 pKa = 11.84QVLFSWYY210 pKa = 9.8VAGGFQGITGFEE222 pKa = 4.36KK223 pKa = 10.43ILTGSGDD230 pKa = 3.56DD231 pKa = 4.87EE232 pKa = 4.47IRR234 pKa = 11.84DD235 pKa = 4.09MIGVEE240 pKa = 4.42YY241 pKa = 10.09IDD243 pKa = 4.27SGSGNDD249 pKa = 3.53LVYY252 pKa = 11.01AGGIPQVLIGGGGSDD267 pKa = 3.69TLILATLFSNPHH279 pKa = 6.93GITLDD284 pKa = 3.44AAQTSGVNGLFGNSTGHH301 pKa = 6.52VIRR304 pKa = 11.84GFEE307 pKa = 4.11NYY309 pKa = 10.0VLTFRR314 pKa = 11.84EE315 pKa = 3.95DD316 pKa = 2.91RR317 pKa = 11.84FYY319 pKa = 11.52GSNKK323 pKa = 9.74SEE325 pKa = 3.93EE326 pKa = 4.29VKK328 pKa = 10.76LGNGNDD334 pKa = 3.32IAFGGAGNDD343 pKa = 3.7TLFGQAGDD351 pKa = 4.42DD352 pKa = 3.95FLDD355 pKa = 4.18GGTGNDD361 pKa = 3.2ILRR364 pKa = 11.84GGGGADD370 pKa = 3.43EE371 pKa = 4.32FHH373 pKa = 7.28FDD375 pKa = 3.1IDD377 pKa = 4.05RR378 pKa = 11.84PFGHH382 pKa = 6.91DD383 pKa = 3.13TVHH386 pKa = 6.49GNSAVDD392 pKa = 3.32TFTFDD397 pKa = 3.21TNGMYY402 pKa = 10.26VGQVQINDD410 pKa = 4.14DD411 pKa = 3.71GDD413 pKa = 3.89DD414 pKa = 3.59AVITFLGLAGSSVTVVGAAGKK435 pKa = 8.23LTQEE439 pKa = 5.36DD440 pKa = 3.99INIIGGFGPVMPDD453 pKa = 3.21DD454 pKa = 4.07FLFF457 pKa = 4.25
MM1 pKa = 7.89RR2 pKa = 11.84ISHH5 pKa = 7.57FYY7 pKa = 10.06TDD9 pKa = 3.56ADD11 pKa = 3.77EE12 pKa = 5.84KK13 pKa = 10.4IFGYY17 pKa = 10.09ISYY20 pKa = 10.05GWTFQNGQWVLGLTFRR36 pKa = 11.84DD37 pKa = 4.08DD38 pKa = 4.18EE39 pKa = 4.25IHH41 pKa = 7.3AGGGDD46 pKa = 3.74DD47 pKa = 4.31TIKK50 pKa = 10.99SSGGDD55 pKa = 3.44DD56 pKa = 3.08IVYY59 pKa = 9.26TGAGADD65 pKa = 3.67FVDD68 pKa = 4.16AGEE71 pKa = 4.72GNDD74 pKa = 4.45TIYY77 pKa = 11.45GDD79 pKa = 3.75AGEE82 pKa = 4.05DD83 pKa = 3.48TIYY86 pKa = 11.19GGDD89 pKa = 3.82GNDD92 pKa = 4.73FIYY95 pKa = 10.51GSPQGGLFYY104 pKa = 11.05GGDD107 pKa = 3.79GNDD110 pKa = 4.03FISGEE115 pKa = 4.24GAAHH119 pKa = 6.0VSIFGDD125 pKa = 3.35AGNDD129 pKa = 3.64TLVGGSGSDD138 pKa = 3.42TVIGGIGNDD147 pKa = 3.25TLYY150 pKa = 9.49ITEE153 pKa = 4.86GQDD156 pKa = 3.16NMHH159 pKa = 6.68GGEE162 pKa = 4.26GDD164 pKa = 3.82DD165 pKa = 5.58VFILDD170 pKa = 4.64QSVADD175 pKa = 3.96LGFRR179 pKa = 11.84TSVFDD184 pKa = 3.82YY185 pKa = 10.84FDD187 pKa = 3.73GSEE190 pKa = 4.71GYY192 pKa = 7.8DD193 pKa = 3.51TLDD196 pKa = 3.04MSAASGRR203 pKa = 11.84QVLFSWYY210 pKa = 9.8VAGGFQGITGFEE222 pKa = 4.36KK223 pKa = 10.43ILTGSGDD230 pKa = 3.56DD231 pKa = 4.87EE232 pKa = 4.47IRR234 pKa = 11.84DD235 pKa = 4.09MIGVEE240 pKa = 4.42YY241 pKa = 10.09IDD243 pKa = 4.27SGSGNDD249 pKa = 3.53LVYY252 pKa = 11.01AGGIPQVLIGGGGSDD267 pKa = 3.69TLILATLFSNPHH279 pKa = 6.93GITLDD284 pKa = 3.44AAQTSGVNGLFGNSTGHH301 pKa = 6.52VIRR304 pKa = 11.84GFEE307 pKa = 4.11NYY309 pKa = 10.0VLTFRR314 pKa = 11.84EE315 pKa = 3.95DD316 pKa = 2.91RR317 pKa = 11.84FYY319 pKa = 11.52GSNKK323 pKa = 9.74SEE325 pKa = 3.93EE326 pKa = 4.29VKK328 pKa = 10.76LGNGNDD334 pKa = 3.32IAFGGAGNDD343 pKa = 3.7TLFGQAGDD351 pKa = 4.42DD352 pKa = 3.95FLDD355 pKa = 4.18GGTGNDD361 pKa = 3.2ILRR364 pKa = 11.84GGGGADD370 pKa = 3.43EE371 pKa = 4.32FHH373 pKa = 7.28FDD375 pKa = 3.1IDD377 pKa = 4.05RR378 pKa = 11.84PFGHH382 pKa = 6.91DD383 pKa = 3.13TVHH386 pKa = 6.49GNSAVDD392 pKa = 3.32TFTFDD397 pKa = 3.21TNGMYY402 pKa = 10.26VGQVQINDD410 pKa = 4.14DD411 pKa = 3.71GDD413 pKa = 3.89DD414 pKa = 3.59AVITFLGLAGSSVTVVGAAGKK435 pKa = 8.23LTQEE439 pKa = 5.36DD440 pKa = 3.99INIIGGFGPVMPDD453 pKa = 3.21DD454 pKa = 4.07FLFF457 pKa = 4.25
Molecular weight: 47.68 kDa
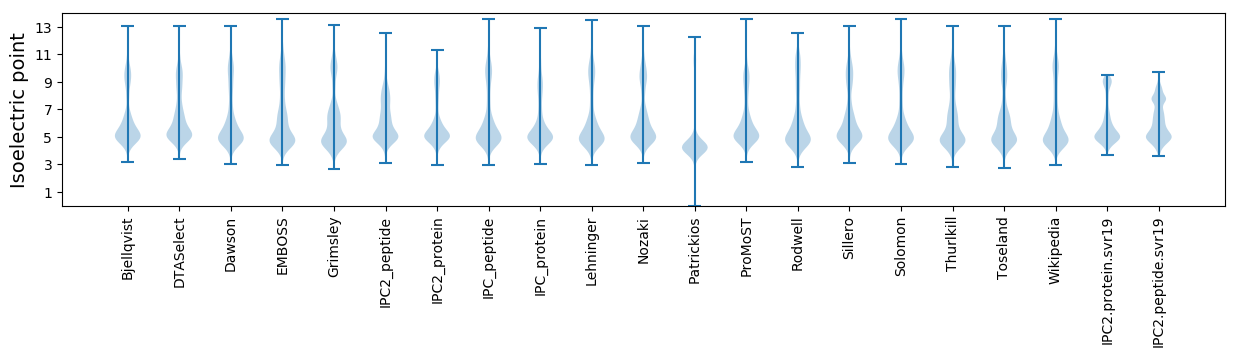
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X7BQG0|A0A1X7BQG0_9RHOB Flagellar biosynthetic protein FlhB OS=Roseovarius aestuarii OX=475083 GN=flhB PE=3 SV=1
MM1 pKa = 7.84RR2 pKa = 11.84GALNAWVVVRR12 pKa = 11.84GALNAWVVVRR22 pKa = 11.84GALNAWVVVRR32 pKa = 11.84GALNAWVVVRR42 pKa = 11.84GTLNAWVVVRR52 pKa = 11.84GTLNALVVVRR62 pKa = 11.84GTLNALVVMRR72 pKa = 11.84GTLNTWVVMRR82 pKa = 11.84GTLNAWVVVRR92 pKa = 11.84GTLNALVVVRR102 pKa = 11.84GTLNALVVVRR112 pKa = 11.84GALNAWVVVRR122 pKa = 11.84GTLNALVVVRR132 pKa = 11.84GALNTWVVMRR142 pKa = 11.84GALNTWVVMRR152 pKa = 11.84GTLNALVVVRR162 pKa = 11.84GTLNALVVVRR172 pKa = 11.84GTLNALVVVRR182 pKa = 11.84GALNAWVVVRR192 pKa = 11.84GTLNALVVVRR202 pKa = 11.84GTLNALVVVRR212 pKa = 11.84GALNTWVVVRR222 pKa = 11.84GALNTWVVVRR232 pKa = 11.84GANARR237 pKa = 11.84FQFDD241 pKa = 3.7LFSWQFRR248 pKa = 11.84NN249 pKa = 3.75
MM1 pKa = 7.84RR2 pKa = 11.84GALNAWVVVRR12 pKa = 11.84GALNAWVVVRR22 pKa = 11.84GALNAWVVVRR32 pKa = 11.84GALNAWVVVRR42 pKa = 11.84GTLNAWVVVRR52 pKa = 11.84GTLNALVVVRR62 pKa = 11.84GTLNALVVMRR72 pKa = 11.84GTLNTWVVMRR82 pKa = 11.84GTLNAWVVVRR92 pKa = 11.84GTLNALVVVRR102 pKa = 11.84GTLNALVVVRR112 pKa = 11.84GALNAWVVVRR122 pKa = 11.84GTLNALVVVRR132 pKa = 11.84GALNTWVVMRR142 pKa = 11.84GALNTWVVMRR152 pKa = 11.84GTLNALVVVRR162 pKa = 11.84GTLNALVVVRR172 pKa = 11.84GTLNALVVVRR182 pKa = 11.84GALNAWVVVRR192 pKa = 11.84GTLNALVVVRR202 pKa = 11.84GTLNALVVVRR212 pKa = 11.84GALNTWVVVRR222 pKa = 11.84GALNTWVVVRR232 pKa = 11.84GANARR237 pKa = 11.84FQFDD241 pKa = 3.7LFSWQFRR248 pKa = 11.84NN249 pKa = 3.75
Molecular weight: 26.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1411738 |
32 |
1938 |
307.3 |
33.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.25 ± 0.041 | 1.01 ± 0.012 |
6.206 ± 0.03 | 5.96 ± 0.029 |
3.872 ± 0.025 | 8.384 ± 0.038 |
2.191 ± 0.016 | 5.52 ± 0.024 |
3.531 ± 0.026 | 9.889 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.801 ± 0.017 | 2.951 ± 0.018 |
4.866 ± 0.025 | 3.305 ± 0.018 |
6.377 ± 0.03 | 5.543 ± 0.023 |
5.48 ± 0.023 | 7.082 ± 0.026 |
1.413 ± 0.016 | 2.368 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
