
Hubei permutotetra-like virus 11
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
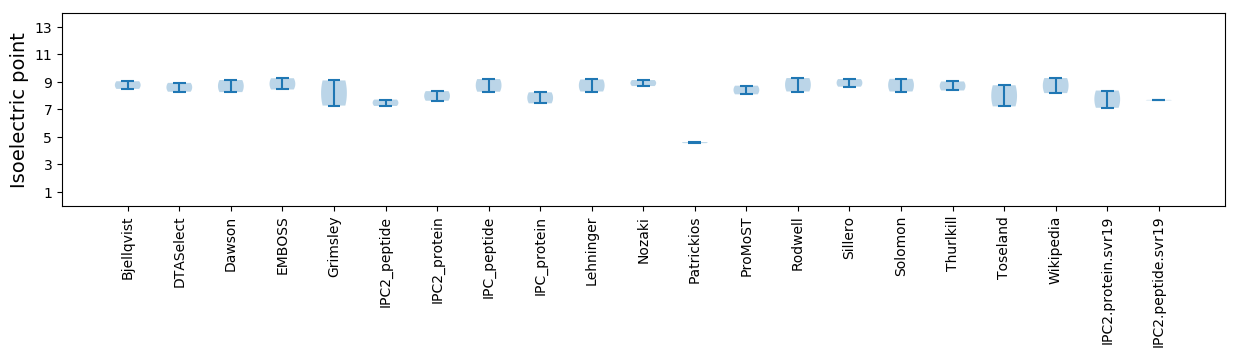
Average proteome isoelectric point is 7.75
Get precalculated fractions of proteins
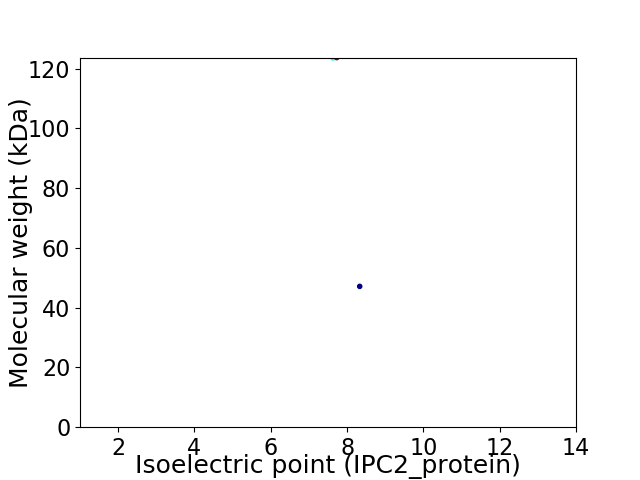
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KHV9|A0A1L3KHV9_9VIRU RdRp OS=Hubei permutotetra-like virus 11 OX=1923075 PE=4 SV=1
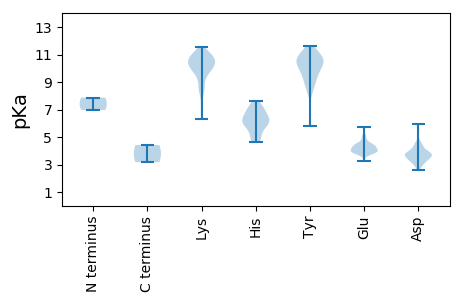
MM1 pKa = 7.87DD2 pKa = 5.59ASNPVISSDD11 pKa = 3.07RR12 pKa = 11.84ATIQSQLEE20 pKa = 4.24KK21 pKa = 10.89VRR23 pKa = 11.84GTRR26 pKa = 11.84RR27 pKa = 11.84QDD29 pKa = 3.01LGYY32 pKa = 10.39LQGLASTTQRR42 pKa = 11.84IQLPTNINIPLLEE55 pKa = 3.86RR56 pKa = 11.84TGIVEE61 pKa = 3.88KK62 pKa = 10.87FKK64 pKa = 10.57TNRR67 pKa = 11.84PEE69 pKa = 3.86VPTTKK74 pKa = 10.51DD75 pKa = 2.78IPLDD79 pKa = 3.66DD80 pKa = 5.32KK81 pKa = 11.54IFMVEE86 pKa = 3.97LNEE89 pKa = 4.53CPVSGILNSNSLPLHH104 pKa = 6.02PSGVNCLGGKK114 pKa = 9.94VNVQRR119 pKa = 11.84KK120 pKa = 9.07VGGIPTAPFGLGDD133 pKa = 3.85IVAVIGDD140 pKa = 3.78DD141 pKa = 3.73VLNTKK146 pKa = 9.62HH147 pKa = 6.52VYY149 pKa = 9.34TSGTWSGFVKK159 pKa = 10.61RR160 pKa = 11.84LNQQMGRR167 pKa = 11.84KK168 pKa = 6.79TVSLRR173 pKa = 11.84KK174 pKa = 8.56TMGNMGKK181 pKa = 9.31EE182 pKa = 4.08LSYY185 pKa = 10.82DD186 pKa = 3.33ACLKK190 pKa = 10.93LLDD193 pKa = 4.48KK194 pKa = 11.2YY195 pKa = 10.35MPRR198 pKa = 11.84KK199 pKa = 9.31PNYY202 pKa = 8.86DD203 pKa = 2.74WPGRR207 pKa = 11.84NEE209 pKa = 3.83ANFYY213 pKa = 11.14EE214 pKa = 4.92DD215 pKa = 3.77LTTKK219 pKa = 10.36IKK221 pKa = 10.06VTMNSSAGAPYY232 pKa = 9.06WRR234 pKa = 11.84NKK236 pKa = 8.5SEE238 pKa = 3.95CMEE241 pKa = 5.17DD242 pKa = 3.5IIDD245 pKa = 3.71VGIPLIVKK253 pKa = 9.89AIKK256 pKa = 10.21EE257 pKa = 3.93NSLDD261 pKa = 3.53KK262 pKa = 11.06LYY264 pKa = 10.55MEE266 pKa = 4.61NPEE269 pKa = 4.11MFLCEE274 pKa = 4.4VKK276 pKa = 10.93NKK278 pKa = 7.46TDD280 pKa = 3.15RR281 pKa = 11.84YY282 pKa = 9.33EE283 pKa = 3.91VSKK286 pKa = 11.19LKK288 pKa = 10.91DD289 pKa = 2.85KK290 pKa = 9.98TRR292 pKa = 11.84PYY294 pKa = 9.81TCVPAHH300 pKa = 6.06WAFLFSMLTQGFQEE314 pKa = 4.2TLLTFDD320 pKa = 4.55QDD322 pKa = 3.8PSSSNAYY329 pKa = 9.88GFSSSNGGLKK339 pKa = 10.48RR340 pKa = 11.84MVEE343 pKa = 3.77WMYY346 pKa = 10.09TADD349 pKa = 3.65RR350 pKa = 11.84RR351 pKa = 11.84GKK353 pKa = 8.35VVCYY357 pKa = 10.56GDD359 pKa = 3.91DD360 pKa = 3.3ACIVIKK366 pKa = 10.68KK367 pKa = 9.45GDD369 pKa = 3.66KK370 pKa = 9.88IYY372 pKa = 10.82RR373 pKa = 11.84IDD375 pKa = 3.99PDD377 pKa = 4.55FKK379 pKa = 11.53QMDD382 pKa = 3.82GSLSQEE388 pKa = 4.41DD389 pKa = 3.64IEE391 pKa = 5.04LTVKK395 pKa = 9.68WVMAHH400 pKa = 5.89FEE402 pKa = 4.11KK403 pKa = 11.0DD404 pKa = 2.91AGKK407 pKa = 10.42QSLFWRR413 pKa = 11.84AVAGVWALMSHH424 pKa = 6.02NPVFVIDD431 pKa = 4.17GKK433 pKa = 8.28TVYY436 pKa = 10.22RR437 pKa = 11.84KK438 pKa = 9.95RR439 pKa = 11.84NPSGLMTGVPGTTLFDD455 pKa = 3.7TVKK458 pKa = 10.97SVMVWNKK465 pKa = 10.32LLDD468 pKa = 3.59VCATGQGDD476 pKa = 3.7ILDD479 pKa = 4.48EE480 pKa = 4.22KK481 pKa = 10.75WVTNWMLQNGLVVKK495 pKa = 10.43EE496 pKa = 4.43GTWKK500 pKa = 10.47PEE502 pKa = 3.96EE503 pKa = 4.24LPKK506 pKa = 10.78QLDD509 pKa = 3.37HH510 pKa = 6.98GDD512 pKa = 3.42LVTTHH517 pKa = 7.02KK518 pKa = 10.74FLGVQIKK525 pKa = 10.23KK526 pKa = 10.23IIWKK530 pKa = 8.68GTEE533 pKa = 3.87QFVPTIPYY541 pKa = 9.2EE542 pKa = 4.31DD543 pKa = 3.87AVEE546 pKa = 3.94MLVIQKK552 pKa = 10.37DD553 pKa = 4.2DD554 pKa = 3.43PWPTNKK560 pKa = 9.44VSNTMKK566 pKa = 10.62ARR568 pKa = 11.84TLFDD572 pKa = 3.36RR573 pKa = 11.84MRR575 pKa = 11.84GLMITMGFNHH585 pKa = 7.12PAIVDD590 pKa = 4.42CIHH593 pKa = 6.41NVVNNLPPEE602 pKa = 4.52VILMQVQNGVGEE614 pKa = 4.4KK615 pKa = 9.32PDD617 pKa = 4.86HH618 pKa = 6.16ITLQEE623 pKa = 3.78FNYY626 pKa = 9.17PDD628 pKa = 3.39SSGFPSVDD636 pKa = 3.73FCISVYY642 pKa = 11.23ADD644 pKa = 3.24GTYY647 pKa = 10.56DD648 pKa = 3.71EE649 pKa = 4.9PWIKK653 pKa = 10.6LFPNLDD659 pKa = 3.77DD660 pKa = 5.47KK661 pKa = 11.46ISAFKK666 pKa = 10.51EE667 pKa = 3.79QIKK670 pKa = 10.4QMEE673 pKa = 4.2RR674 pKa = 11.84KK675 pKa = 9.13YY676 pKa = 10.92RR677 pKa = 11.84EE678 pKa = 3.92LLGVQDD684 pKa = 3.83PQYY687 pKa = 11.64VEE689 pKa = 5.61IEE691 pKa = 4.29DD692 pKa = 3.91KK693 pKa = 10.87PLNQEE698 pKa = 4.04YY699 pKa = 10.74QLVEE703 pKa = 3.86AKK705 pKa = 10.11TPVKK709 pKa = 9.9PPKK712 pKa = 10.05FEE714 pKa = 4.45SVNKK718 pKa = 9.63RR719 pKa = 11.84SKK721 pKa = 9.52VCEE724 pKa = 3.85VHH726 pKa = 7.63DD727 pKa = 5.96DD728 pKa = 4.43RR729 pKa = 11.84IQDD732 pKa = 3.25RR733 pKa = 11.84KK734 pKa = 10.28YY735 pKa = 11.06VPNLGQSVRR744 pKa = 11.84IFLQKK749 pKa = 10.72GYY751 pKa = 10.34GLAPVGVVAAGLGVSARR768 pKa = 11.84RR769 pKa = 11.84LYY771 pKa = 10.8RR772 pKa = 11.84EE773 pKa = 3.81ASNWGYY779 pKa = 11.65YY780 pKa = 5.82MTGWTVDD787 pKa = 4.53DD788 pKa = 3.92ICSLRR793 pKa = 11.84PIITAFKK800 pKa = 8.78THH802 pKa = 6.19QEE804 pKa = 4.27DD805 pKa = 4.09IFDD808 pKa = 4.21DD809 pKa = 4.02MEE811 pKa = 4.49GKK813 pKa = 8.26KK814 pKa = 8.44TLINKK819 pKa = 7.42STEE822 pKa = 3.41IRR824 pKa = 11.84QEE826 pKa = 3.95SLQKK830 pKa = 10.13SGEE833 pKa = 4.4VVRR836 pKa = 11.84SAPDD840 pKa = 3.31LVLLRR845 pKa = 11.84KK846 pKa = 10.24DD847 pKa = 2.9ILLFNQSAPVSQVDD861 pKa = 3.56NADD864 pKa = 3.54EE865 pKa = 4.14AFKK868 pKa = 10.7ILNLMVSRR876 pKa = 11.84EE877 pKa = 3.81GLQYY881 pKa = 11.02TFTTKK886 pKa = 9.9VDD888 pKa = 3.35ATKK891 pKa = 10.28VNSVGVVMVARR902 pKa = 11.84HH903 pKa = 5.8SSTGVPSGWTPVAEE917 pKa = 3.91AWSLNKK923 pKa = 10.04KK924 pKa = 8.68LAKK927 pKa = 10.13EE928 pKa = 4.52FIARR932 pKa = 11.84SILEE936 pKa = 4.03MNGIEE941 pKa = 4.16CEE943 pKa = 3.85EE944 pKa = 4.5SKK946 pKa = 11.03FSVTYY951 pKa = 9.66VPPILEE957 pKa = 4.51EE958 pKa = 4.06SLNWADD964 pKa = 3.65EE965 pKa = 4.42VEE967 pKa = 4.43NTFNPRR973 pKa = 11.84QIPRR977 pKa = 11.84VIDD980 pKa = 3.67EE981 pKa = 4.61KK982 pKa = 11.25NSNVLPDD989 pKa = 3.74LDD991 pKa = 4.21VYY993 pKa = 11.17QEE995 pKa = 4.13LEE997 pKa = 3.9KK998 pKa = 10.84LRR1000 pKa = 11.84PYY1002 pKa = 10.17IPKK1005 pKa = 10.16KK1006 pKa = 9.38FLGYY1010 pKa = 10.61AEE1012 pKa = 4.39SLFVKK1017 pKa = 9.95PNPQEE1022 pKa = 3.89HH1023 pKa = 6.45LKK1025 pKa = 10.81AALEE1029 pKa = 3.99MLDD1032 pKa = 3.84KK1033 pKa = 10.31IDD1035 pKa = 3.72RR1036 pKa = 11.84QVHH1039 pKa = 5.42HH1040 pKa = 7.03WSSSDD1045 pKa = 3.16SDD1047 pKa = 3.76TSGVGSSSPSTPSKK1061 pKa = 10.46RR1062 pKa = 11.84SRR1064 pKa = 11.84LTPARR1069 pKa = 11.84RR1070 pKa = 11.84TKK1072 pKa = 10.81LNRR1075 pKa = 11.84KK1076 pKa = 6.29LTEE1079 pKa = 3.29RR1080 pKa = 11.84RR1081 pKa = 11.84RR1082 pKa = 11.84RR1083 pKa = 11.84KK1084 pKa = 9.55KK1085 pKa = 10.56ASSTTKK1091 pKa = 10.12PP1092 pKa = 3.18
MM1 pKa = 7.87DD2 pKa = 5.59ASNPVISSDD11 pKa = 3.07RR12 pKa = 11.84ATIQSQLEE20 pKa = 4.24KK21 pKa = 10.89VRR23 pKa = 11.84GTRR26 pKa = 11.84RR27 pKa = 11.84QDD29 pKa = 3.01LGYY32 pKa = 10.39LQGLASTTQRR42 pKa = 11.84IQLPTNINIPLLEE55 pKa = 3.86RR56 pKa = 11.84TGIVEE61 pKa = 3.88KK62 pKa = 10.87FKK64 pKa = 10.57TNRR67 pKa = 11.84PEE69 pKa = 3.86VPTTKK74 pKa = 10.51DD75 pKa = 2.78IPLDD79 pKa = 3.66DD80 pKa = 5.32KK81 pKa = 11.54IFMVEE86 pKa = 3.97LNEE89 pKa = 4.53CPVSGILNSNSLPLHH104 pKa = 6.02PSGVNCLGGKK114 pKa = 9.94VNVQRR119 pKa = 11.84KK120 pKa = 9.07VGGIPTAPFGLGDD133 pKa = 3.85IVAVIGDD140 pKa = 3.78DD141 pKa = 3.73VLNTKK146 pKa = 9.62HH147 pKa = 6.52VYY149 pKa = 9.34TSGTWSGFVKK159 pKa = 10.61RR160 pKa = 11.84LNQQMGRR167 pKa = 11.84KK168 pKa = 6.79TVSLRR173 pKa = 11.84KK174 pKa = 8.56TMGNMGKK181 pKa = 9.31EE182 pKa = 4.08LSYY185 pKa = 10.82DD186 pKa = 3.33ACLKK190 pKa = 10.93LLDD193 pKa = 4.48KK194 pKa = 11.2YY195 pKa = 10.35MPRR198 pKa = 11.84KK199 pKa = 9.31PNYY202 pKa = 8.86DD203 pKa = 2.74WPGRR207 pKa = 11.84NEE209 pKa = 3.83ANFYY213 pKa = 11.14EE214 pKa = 4.92DD215 pKa = 3.77LTTKK219 pKa = 10.36IKK221 pKa = 10.06VTMNSSAGAPYY232 pKa = 9.06WRR234 pKa = 11.84NKK236 pKa = 8.5SEE238 pKa = 3.95CMEE241 pKa = 5.17DD242 pKa = 3.5IIDD245 pKa = 3.71VGIPLIVKK253 pKa = 9.89AIKK256 pKa = 10.21EE257 pKa = 3.93NSLDD261 pKa = 3.53KK262 pKa = 11.06LYY264 pKa = 10.55MEE266 pKa = 4.61NPEE269 pKa = 4.11MFLCEE274 pKa = 4.4VKK276 pKa = 10.93NKK278 pKa = 7.46TDD280 pKa = 3.15RR281 pKa = 11.84YY282 pKa = 9.33EE283 pKa = 3.91VSKK286 pKa = 11.19LKK288 pKa = 10.91DD289 pKa = 2.85KK290 pKa = 9.98TRR292 pKa = 11.84PYY294 pKa = 9.81TCVPAHH300 pKa = 6.06WAFLFSMLTQGFQEE314 pKa = 4.2TLLTFDD320 pKa = 4.55QDD322 pKa = 3.8PSSSNAYY329 pKa = 9.88GFSSSNGGLKK339 pKa = 10.48RR340 pKa = 11.84MVEE343 pKa = 3.77WMYY346 pKa = 10.09TADD349 pKa = 3.65RR350 pKa = 11.84RR351 pKa = 11.84GKK353 pKa = 8.35VVCYY357 pKa = 10.56GDD359 pKa = 3.91DD360 pKa = 3.3ACIVIKK366 pKa = 10.68KK367 pKa = 9.45GDD369 pKa = 3.66KK370 pKa = 9.88IYY372 pKa = 10.82RR373 pKa = 11.84IDD375 pKa = 3.99PDD377 pKa = 4.55FKK379 pKa = 11.53QMDD382 pKa = 3.82GSLSQEE388 pKa = 4.41DD389 pKa = 3.64IEE391 pKa = 5.04LTVKK395 pKa = 9.68WVMAHH400 pKa = 5.89FEE402 pKa = 4.11KK403 pKa = 11.0DD404 pKa = 2.91AGKK407 pKa = 10.42QSLFWRR413 pKa = 11.84AVAGVWALMSHH424 pKa = 6.02NPVFVIDD431 pKa = 4.17GKK433 pKa = 8.28TVYY436 pKa = 10.22RR437 pKa = 11.84KK438 pKa = 9.95RR439 pKa = 11.84NPSGLMTGVPGTTLFDD455 pKa = 3.7TVKK458 pKa = 10.97SVMVWNKK465 pKa = 10.32LLDD468 pKa = 3.59VCATGQGDD476 pKa = 3.7ILDD479 pKa = 4.48EE480 pKa = 4.22KK481 pKa = 10.75WVTNWMLQNGLVVKK495 pKa = 10.43EE496 pKa = 4.43GTWKK500 pKa = 10.47PEE502 pKa = 3.96EE503 pKa = 4.24LPKK506 pKa = 10.78QLDD509 pKa = 3.37HH510 pKa = 6.98GDD512 pKa = 3.42LVTTHH517 pKa = 7.02KK518 pKa = 10.74FLGVQIKK525 pKa = 10.23KK526 pKa = 10.23IIWKK530 pKa = 8.68GTEE533 pKa = 3.87QFVPTIPYY541 pKa = 9.2EE542 pKa = 4.31DD543 pKa = 3.87AVEE546 pKa = 3.94MLVIQKK552 pKa = 10.37DD553 pKa = 4.2DD554 pKa = 3.43PWPTNKK560 pKa = 9.44VSNTMKK566 pKa = 10.62ARR568 pKa = 11.84TLFDD572 pKa = 3.36RR573 pKa = 11.84MRR575 pKa = 11.84GLMITMGFNHH585 pKa = 7.12PAIVDD590 pKa = 4.42CIHH593 pKa = 6.41NVVNNLPPEE602 pKa = 4.52VILMQVQNGVGEE614 pKa = 4.4KK615 pKa = 9.32PDD617 pKa = 4.86HH618 pKa = 6.16ITLQEE623 pKa = 3.78FNYY626 pKa = 9.17PDD628 pKa = 3.39SSGFPSVDD636 pKa = 3.73FCISVYY642 pKa = 11.23ADD644 pKa = 3.24GTYY647 pKa = 10.56DD648 pKa = 3.71EE649 pKa = 4.9PWIKK653 pKa = 10.6LFPNLDD659 pKa = 3.77DD660 pKa = 5.47KK661 pKa = 11.46ISAFKK666 pKa = 10.51EE667 pKa = 3.79QIKK670 pKa = 10.4QMEE673 pKa = 4.2RR674 pKa = 11.84KK675 pKa = 9.13YY676 pKa = 10.92RR677 pKa = 11.84EE678 pKa = 3.92LLGVQDD684 pKa = 3.83PQYY687 pKa = 11.64VEE689 pKa = 5.61IEE691 pKa = 4.29DD692 pKa = 3.91KK693 pKa = 10.87PLNQEE698 pKa = 4.04YY699 pKa = 10.74QLVEE703 pKa = 3.86AKK705 pKa = 10.11TPVKK709 pKa = 9.9PPKK712 pKa = 10.05FEE714 pKa = 4.45SVNKK718 pKa = 9.63RR719 pKa = 11.84SKK721 pKa = 9.52VCEE724 pKa = 3.85VHH726 pKa = 7.63DD727 pKa = 5.96DD728 pKa = 4.43RR729 pKa = 11.84IQDD732 pKa = 3.25RR733 pKa = 11.84KK734 pKa = 10.28YY735 pKa = 11.06VPNLGQSVRR744 pKa = 11.84IFLQKK749 pKa = 10.72GYY751 pKa = 10.34GLAPVGVVAAGLGVSARR768 pKa = 11.84RR769 pKa = 11.84LYY771 pKa = 10.8RR772 pKa = 11.84EE773 pKa = 3.81ASNWGYY779 pKa = 11.65YY780 pKa = 5.82MTGWTVDD787 pKa = 4.53DD788 pKa = 3.92ICSLRR793 pKa = 11.84PIITAFKK800 pKa = 8.78THH802 pKa = 6.19QEE804 pKa = 4.27DD805 pKa = 4.09IFDD808 pKa = 4.21DD809 pKa = 4.02MEE811 pKa = 4.49GKK813 pKa = 8.26KK814 pKa = 8.44TLINKK819 pKa = 7.42STEE822 pKa = 3.41IRR824 pKa = 11.84QEE826 pKa = 3.95SLQKK830 pKa = 10.13SGEE833 pKa = 4.4VVRR836 pKa = 11.84SAPDD840 pKa = 3.31LVLLRR845 pKa = 11.84KK846 pKa = 10.24DD847 pKa = 2.9ILLFNQSAPVSQVDD861 pKa = 3.56NADD864 pKa = 3.54EE865 pKa = 4.14AFKK868 pKa = 10.7ILNLMVSRR876 pKa = 11.84EE877 pKa = 3.81GLQYY881 pKa = 11.02TFTTKK886 pKa = 9.9VDD888 pKa = 3.35ATKK891 pKa = 10.28VNSVGVVMVARR902 pKa = 11.84HH903 pKa = 5.8SSTGVPSGWTPVAEE917 pKa = 3.91AWSLNKK923 pKa = 10.04KK924 pKa = 8.68LAKK927 pKa = 10.13EE928 pKa = 4.52FIARR932 pKa = 11.84SILEE936 pKa = 4.03MNGIEE941 pKa = 4.16CEE943 pKa = 3.85EE944 pKa = 4.5SKK946 pKa = 11.03FSVTYY951 pKa = 9.66VPPILEE957 pKa = 4.51EE958 pKa = 4.06SLNWADD964 pKa = 3.65EE965 pKa = 4.42VEE967 pKa = 4.43NTFNPRR973 pKa = 11.84QIPRR977 pKa = 11.84VIDD980 pKa = 3.67EE981 pKa = 4.61KK982 pKa = 11.25NSNVLPDD989 pKa = 3.74LDD991 pKa = 4.21VYY993 pKa = 11.17QEE995 pKa = 4.13LEE997 pKa = 3.9KK998 pKa = 10.84LRR1000 pKa = 11.84PYY1002 pKa = 10.17IPKK1005 pKa = 10.16KK1006 pKa = 9.38FLGYY1010 pKa = 10.61AEE1012 pKa = 4.39SLFVKK1017 pKa = 9.95PNPQEE1022 pKa = 3.89HH1023 pKa = 6.45LKK1025 pKa = 10.81AALEE1029 pKa = 3.99MLDD1032 pKa = 3.84KK1033 pKa = 10.31IDD1035 pKa = 3.72RR1036 pKa = 11.84QVHH1039 pKa = 5.42HH1040 pKa = 7.03WSSSDD1045 pKa = 3.16SDD1047 pKa = 3.76TSGVGSSSPSTPSKK1061 pKa = 10.46RR1062 pKa = 11.84SRR1064 pKa = 11.84LTPARR1069 pKa = 11.84RR1070 pKa = 11.84TKK1072 pKa = 10.81LNRR1075 pKa = 11.84KK1076 pKa = 6.29LTEE1079 pKa = 3.29RR1080 pKa = 11.84RR1081 pKa = 11.84RR1082 pKa = 11.84RR1083 pKa = 11.84KK1084 pKa = 9.55KK1085 pKa = 10.56ASSTTKK1091 pKa = 10.12PP1092 pKa = 3.18
Molecular weight: 123.56 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KHV9|A0A1L3KHV9_9VIRU RdRp OS=Hubei permutotetra-like virus 11 OX=1923075 PE=4 SV=1
MM1 pKa = 6.97VGSVEE6 pKa = 5.12CEE8 pKa = 3.8YY9 pKa = 10.96CKK11 pKa = 10.79KK12 pKa = 10.45KK13 pKa = 10.78FKK15 pKa = 10.77SKK17 pKa = 9.95RR18 pKa = 11.84AKK20 pKa = 8.4NQHH23 pKa = 4.69VSMVHH28 pKa = 6.94KK29 pKa = 10.14INEE32 pKa = 4.1KK33 pKa = 8.71MAPPAVAKK41 pKa = 9.73TGRR44 pKa = 11.84TRR46 pKa = 11.84GSLRR50 pKa = 11.84LRR52 pKa = 11.84RR53 pKa = 11.84TRR55 pKa = 11.84TSNLPGFDD63 pKa = 3.31IAPSRR68 pKa = 11.84VPTVRR73 pKa = 11.84GGMINISGEE82 pKa = 3.97DD83 pKa = 3.28RR84 pKa = 11.84IGAFDD89 pKa = 4.46LKK91 pKa = 10.72SGKK94 pKa = 10.05AVFMSVDD101 pKa = 3.57ISPSMSARR109 pKa = 11.84LTTIARR115 pKa = 11.84AYY117 pKa = 10.39QRR119 pKa = 11.84IKK121 pKa = 9.44WNAVKK126 pKa = 10.35IVVTPQASAMTNGGYY141 pKa = 10.01VCGFIADD148 pKa = 3.86PSDD151 pKa = 3.57RR152 pKa = 11.84AVTASDD158 pKa = 3.99LSASQGAQTKK168 pKa = 9.89KK169 pKa = 10.01FYY171 pKa = 8.59EE172 pKa = 4.21TAVVYY177 pKa = 8.45MPRR180 pKa = 11.84KK181 pKa = 8.11TDD183 pKa = 3.81LLYY186 pKa = 10.66TSAGEE191 pKa = 4.05DD192 pKa = 3.19PRR194 pKa = 11.84LFMPASFWIISEE206 pKa = 4.27GLPSSNLTMIVSVVWDD222 pKa = 3.72VTLSQPTLEE231 pKa = 4.24NSHH234 pKa = 6.41NNSFLLVGEE243 pKa = 4.7IVPNPANYY251 pKa = 9.85NLRR254 pKa = 11.84YY255 pKa = 9.52SPPGGDD261 pKa = 3.67PQDD264 pKa = 4.29DD265 pKa = 3.76ASSIIPAVLRR275 pKa = 11.84EE276 pKa = 4.23TPGYY280 pKa = 10.5HH281 pKa = 5.91YY282 pKa = 10.72FRR284 pKa = 11.84VPTFTIEE291 pKa = 4.02YY292 pKa = 10.33SEE294 pKa = 4.46GTGDD298 pKa = 3.17TGTIQAHH305 pKa = 4.63FVVYY309 pKa = 8.48HH310 pKa = 5.01TTDD313 pKa = 2.63KK314 pKa = 11.14KK315 pKa = 11.08LYY317 pKa = 8.26YY318 pKa = 10.02SSNGRR323 pKa = 11.84DD324 pKa = 3.2VEE326 pKa = 4.4TTMWQGNVDD335 pKa = 3.31AHH337 pKa = 5.12QTLVPCGTFMKK348 pKa = 10.81YY349 pKa = 9.92VGQEE353 pKa = 3.6NSCRR357 pKa = 11.84DD358 pKa = 3.8TKK360 pKa = 11.36LLTPKK365 pKa = 10.21LLSEE369 pKa = 4.55TSPGSTSSSTNLSAKK384 pKa = 6.77MQKK387 pKa = 10.52LEE389 pKa = 4.09TLFLEE394 pKa = 5.76LKK396 pKa = 10.53QNYY399 pKa = 9.19QKK401 pKa = 10.99DD402 pKa = 4.19SKK404 pKa = 11.0PLMEE408 pKa = 5.02EE409 pKa = 4.28SMILKK414 pKa = 10.01PNLVEE419 pKa = 4.87KK420 pKa = 11.06LEE422 pKa = 3.91QLEE425 pKa = 4.25LL426 pKa = 4.42
MM1 pKa = 6.97VGSVEE6 pKa = 5.12CEE8 pKa = 3.8YY9 pKa = 10.96CKK11 pKa = 10.79KK12 pKa = 10.45KK13 pKa = 10.78FKK15 pKa = 10.77SKK17 pKa = 9.95RR18 pKa = 11.84AKK20 pKa = 8.4NQHH23 pKa = 4.69VSMVHH28 pKa = 6.94KK29 pKa = 10.14INEE32 pKa = 4.1KK33 pKa = 8.71MAPPAVAKK41 pKa = 9.73TGRR44 pKa = 11.84TRR46 pKa = 11.84GSLRR50 pKa = 11.84LRR52 pKa = 11.84RR53 pKa = 11.84TRR55 pKa = 11.84TSNLPGFDD63 pKa = 3.31IAPSRR68 pKa = 11.84VPTVRR73 pKa = 11.84GGMINISGEE82 pKa = 3.97DD83 pKa = 3.28RR84 pKa = 11.84IGAFDD89 pKa = 4.46LKK91 pKa = 10.72SGKK94 pKa = 10.05AVFMSVDD101 pKa = 3.57ISPSMSARR109 pKa = 11.84LTTIARR115 pKa = 11.84AYY117 pKa = 10.39QRR119 pKa = 11.84IKK121 pKa = 9.44WNAVKK126 pKa = 10.35IVVTPQASAMTNGGYY141 pKa = 10.01VCGFIADD148 pKa = 3.86PSDD151 pKa = 3.57RR152 pKa = 11.84AVTASDD158 pKa = 3.99LSASQGAQTKK168 pKa = 9.89KK169 pKa = 10.01FYY171 pKa = 8.59EE172 pKa = 4.21TAVVYY177 pKa = 8.45MPRR180 pKa = 11.84KK181 pKa = 8.11TDD183 pKa = 3.81LLYY186 pKa = 10.66TSAGEE191 pKa = 4.05DD192 pKa = 3.19PRR194 pKa = 11.84LFMPASFWIISEE206 pKa = 4.27GLPSSNLTMIVSVVWDD222 pKa = 3.72VTLSQPTLEE231 pKa = 4.24NSHH234 pKa = 6.41NNSFLLVGEE243 pKa = 4.7IVPNPANYY251 pKa = 9.85NLRR254 pKa = 11.84YY255 pKa = 9.52SPPGGDD261 pKa = 3.67PQDD264 pKa = 4.29DD265 pKa = 3.76ASSIIPAVLRR275 pKa = 11.84EE276 pKa = 4.23TPGYY280 pKa = 10.5HH281 pKa = 5.91YY282 pKa = 10.72FRR284 pKa = 11.84VPTFTIEE291 pKa = 4.02YY292 pKa = 10.33SEE294 pKa = 4.46GTGDD298 pKa = 3.17TGTIQAHH305 pKa = 4.63FVVYY309 pKa = 8.48HH310 pKa = 5.01TTDD313 pKa = 2.63KK314 pKa = 11.14KK315 pKa = 11.08LYY317 pKa = 8.26YY318 pKa = 10.02SSNGRR323 pKa = 11.84DD324 pKa = 3.2VEE326 pKa = 4.4TTMWQGNVDD335 pKa = 3.31AHH337 pKa = 5.12QTLVPCGTFMKK348 pKa = 10.81YY349 pKa = 9.92VGQEE353 pKa = 3.6NSCRR357 pKa = 11.84DD358 pKa = 3.8TKK360 pKa = 11.36LLTPKK365 pKa = 10.21LLSEE369 pKa = 4.55TSPGSTSSSTNLSAKK384 pKa = 6.77MQKK387 pKa = 10.52LEE389 pKa = 4.09TLFLEE394 pKa = 5.76LKK396 pKa = 10.53QNYY399 pKa = 9.19QKK401 pKa = 10.99DD402 pKa = 4.19SKK404 pKa = 11.0PLMEE408 pKa = 5.02EE409 pKa = 4.28SMILKK414 pKa = 10.01PNLVEE419 pKa = 4.87KK420 pKa = 11.06LEE422 pKa = 3.91QLEE425 pKa = 4.25LL426 pKa = 4.42
Molecular weight: 47.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1518 |
426 |
1092 |
759.0 |
85.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.007 ± 0.676 | 1.252 ± 0.04 |
5.929 ± 0.746 | 5.797 ± 0.321 |
3.36 ± 0.037 | 6.192 ± 0.074 |
1.515 ± 0.065 | 5.072 ± 0.311 |
7.773 ± 0.61 | 8.3 ± 0.162 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.096 ± 0.216 | 4.809 ± 0.177 |
5.863 ± 0.122 | 3.821 ± 0.152 |
5.007 ± 0.039 | 7.642 ± 1.126 |
6.653 ± 0.793 | 8.103 ± 0.419 |
1.647 ± 0.359 | 3.162 ± 0.301 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
