
Dragonfly-associated mastrevirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Mastrevirus
Average proteome isoelectric point is 8.1
Get precalculated fractions of proteins
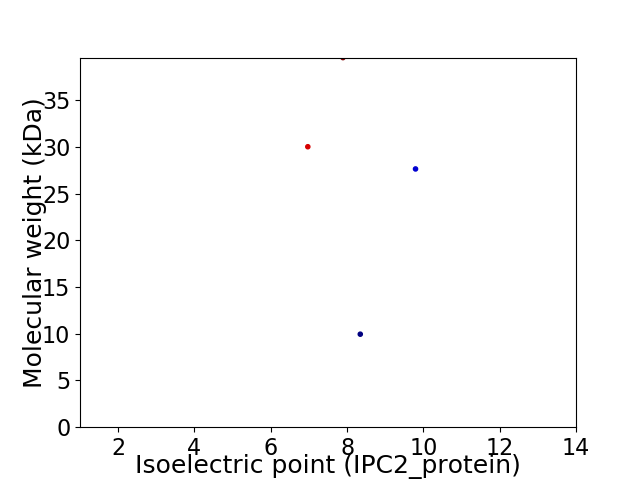
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
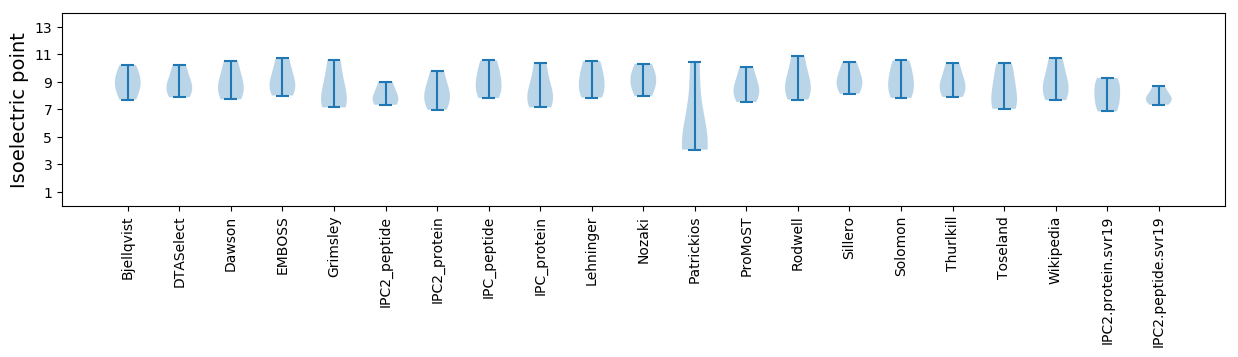
Protein with the lowest isoelectric point:
>tr|K7S1S4|K7S1S4_9GEMI Movement protein OS=Dragonfly-associated mastrevirus OX=1249648 PE=3 SV=1
MM1 pKa = 7.51SSSSAQSGPSHH12 pKa = 6.28FRR14 pKa = 11.84IRR16 pKa = 11.84AQNIFLTYY24 pKa = 9.3PRR26 pKa = 11.84CDD28 pKa = 4.85LDD30 pKa = 4.05PKK32 pKa = 10.82DD33 pKa = 4.04AGEE36 pKa = 4.62IIQSKK41 pKa = 7.32MQSHH45 pKa = 5.89EE46 pKa = 3.69PKK48 pKa = 10.19YY49 pKa = 10.44ILFSRR54 pKa = 11.84EE55 pKa = 3.67LHH57 pKa = 6.66SDD59 pKa = 3.84GEE61 pKa = 4.47YY62 pKa = 10.45HH63 pKa = 7.16LHH65 pKa = 7.17GLLQLSRR72 pKa = 11.84QFSSNNPRR80 pKa = 11.84IFDD83 pKa = 3.5IGAHH87 pKa = 6.05HH88 pKa = 7.22PNIQSAISPKK98 pKa = 9.44SVRR101 pKa = 11.84DD102 pKa = 3.8YY103 pKa = 11.1ILKK106 pKa = 10.5NPITQFCIGTYY117 pKa = 10.2VPAKK121 pKa = 9.94KK122 pKa = 10.02GRR124 pKa = 11.84KK125 pKa = 8.27LGSRR129 pKa = 11.84FEE131 pKa = 4.21EE132 pKa = 4.2NIRR135 pKa = 11.84NNIMRR140 pKa = 11.84SIISTATSKK149 pKa = 10.8EE150 pKa = 4.21PYY152 pKa = 9.9LSMVRR157 pKa = 11.84KK158 pKa = 9.96SFPFEE163 pKa = 3.57WATKK167 pKa = 10.13LSQFEE172 pKa = 4.2YY173 pKa = 10.28SASKK177 pKa = 10.45LFPEE181 pKa = 4.31VTPEE185 pKa = 3.98YY186 pKa = 10.26KK187 pKa = 10.48SPFPTEE193 pKa = 4.2SLICNEE199 pKa = 5.08NIQDD203 pKa = 3.54WVDD206 pKa = 3.08NTLYY210 pKa = 10.33QVSPASYY217 pKa = 10.57CLLHH221 pKa = 6.03PHH223 pKa = 7.23ASATSDD229 pKa = 4.1LLWLQSTSFSLIAPLGGFPSTYY251 pKa = 10.63ADD253 pKa = 3.24QQGQEE258 pKa = 4.07RR259 pKa = 11.84LPGQEE264 pKa = 3.77VV265 pKa = 3.09
MM1 pKa = 7.51SSSSAQSGPSHH12 pKa = 6.28FRR14 pKa = 11.84IRR16 pKa = 11.84AQNIFLTYY24 pKa = 9.3PRR26 pKa = 11.84CDD28 pKa = 4.85LDD30 pKa = 4.05PKK32 pKa = 10.82DD33 pKa = 4.04AGEE36 pKa = 4.62IIQSKK41 pKa = 7.32MQSHH45 pKa = 5.89EE46 pKa = 3.69PKK48 pKa = 10.19YY49 pKa = 10.44ILFSRR54 pKa = 11.84EE55 pKa = 3.67LHH57 pKa = 6.66SDD59 pKa = 3.84GEE61 pKa = 4.47YY62 pKa = 10.45HH63 pKa = 7.16LHH65 pKa = 7.17GLLQLSRR72 pKa = 11.84QFSSNNPRR80 pKa = 11.84IFDD83 pKa = 3.5IGAHH87 pKa = 6.05HH88 pKa = 7.22PNIQSAISPKK98 pKa = 9.44SVRR101 pKa = 11.84DD102 pKa = 3.8YY103 pKa = 11.1ILKK106 pKa = 10.5NPITQFCIGTYY117 pKa = 10.2VPAKK121 pKa = 9.94KK122 pKa = 10.02GRR124 pKa = 11.84KK125 pKa = 8.27LGSRR129 pKa = 11.84FEE131 pKa = 4.21EE132 pKa = 4.2NIRR135 pKa = 11.84NNIMRR140 pKa = 11.84SIISTATSKK149 pKa = 10.8EE150 pKa = 4.21PYY152 pKa = 9.9LSMVRR157 pKa = 11.84KK158 pKa = 9.96SFPFEE163 pKa = 3.57WATKK167 pKa = 10.13LSQFEE172 pKa = 4.2YY173 pKa = 10.28SASKK177 pKa = 10.45LFPEE181 pKa = 4.31VTPEE185 pKa = 3.98YY186 pKa = 10.26KK187 pKa = 10.48SPFPTEE193 pKa = 4.2SLICNEE199 pKa = 5.08NIQDD203 pKa = 3.54WVDD206 pKa = 3.08NTLYY210 pKa = 10.33QVSPASYY217 pKa = 10.57CLLHH221 pKa = 6.03PHH223 pKa = 7.23ASATSDD229 pKa = 4.1LLWLQSTSFSLIAPLGGFPSTYY251 pKa = 10.63ADD253 pKa = 3.24QQGQEE258 pKa = 4.07RR259 pKa = 11.84LPGQEE264 pKa = 3.77VV265 pKa = 3.09
Molecular weight: 30.02 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K7RTG7|K7RTG7_9GEMI Replication-associated protein OS=Dragonfly-associated mastrevirus OX=1249648 PE=3 SV=1
MM1 pKa = 7.44KK2 pKa = 10.14RR3 pKa = 11.84PSEE6 pKa = 3.86GRR8 pKa = 11.84YY9 pKa = 9.25KK10 pKa = 10.66GNVYY14 pKa = 10.41KK15 pKa = 10.48RR16 pKa = 11.84VKK18 pKa = 10.19PGTSGGNTSGGSVVARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84SGWRR41 pKa = 11.84PSPLQFIQYY50 pKa = 7.68TWTTGGSPITVGAGGYY66 pKa = 8.99VALLTSYY73 pKa = 10.66PRR75 pKa = 11.84GSDD78 pKa = 2.79EE79 pKa = 4.74DD80 pKa = 3.61KK81 pKa = 10.81RR82 pKa = 11.84HH83 pKa = 5.45TGEE86 pKa = 3.8TLTYY90 pKa = 10.44KK91 pKa = 10.7VGLDD95 pKa = 3.46LVFNYY100 pKa = 9.64NSKK103 pKa = 9.39MATGRR108 pKa = 11.84IARR111 pKa = 11.84THH113 pKa = 5.27FCVWLVYY120 pKa = 10.39DD121 pKa = 4.23SQPSGTLPAAHH132 pKa = 7.37GIFDD136 pKa = 6.05LIDD139 pKa = 3.56SFTDD143 pKa = 4.54YY144 pKa = 10.59PATWKK149 pKa = 10.34INRR152 pKa = 11.84DD153 pKa = 3.07MGHH156 pKa = 6.44RR157 pKa = 11.84FVIKK161 pKa = 10.17RR162 pKa = 11.84RR163 pKa = 11.84WVNWLEE169 pKa = 3.95ADD171 pKa = 4.24GSISTSNYY179 pKa = 9.46SGAPSAPAKK188 pKa = 10.53SSVVFNKK195 pKa = 9.4FVKK198 pKa = 10.13RR199 pKa = 11.84LGVRR203 pKa = 11.84TEE205 pKa = 3.93WKK207 pKa = 8.31NTTTGGIGDD216 pKa = 3.75VSKK219 pKa = 10.55GALYY223 pKa = 10.2IVVARR228 pKa = 11.84QLYY231 pKa = 10.21DD232 pKa = 3.02IDD234 pKa = 3.42VRR236 pKa = 11.84GRR238 pKa = 11.84LRR240 pKa = 11.84VYY242 pKa = 10.06FKK244 pKa = 11.14SVGNQQ249 pKa = 2.8
MM1 pKa = 7.44KK2 pKa = 10.14RR3 pKa = 11.84PSEE6 pKa = 3.86GRR8 pKa = 11.84YY9 pKa = 9.25KK10 pKa = 10.66GNVYY14 pKa = 10.41KK15 pKa = 10.48RR16 pKa = 11.84VKK18 pKa = 10.19PGTSGGNTSGGSVVARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84SGWRR41 pKa = 11.84PSPLQFIQYY50 pKa = 7.68TWTTGGSPITVGAGGYY66 pKa = 8.99VALLTSYY73 pKa = 10.66PRR75 pKa = 11.84GSDD78 pKa = 2.79EE79 pKa = 4.74DD80 pKa = 3.61KK81 pKa = 10.81RR82 pKa = 11.84HH83 pKa = 5.45TGEE86 pKa = 3.8TLTYY90 pKa = 10.44KK91 pKa = 10.7VGLDD95 pKa = 3.46LVFNYY100 pKa = 9.64NSKK103 pKa = 9.39MATGRR108 pKa = 11.84IARR111 pKa = 11.84THH113 pKa = 5.27FCVWLVYY120 pKa = 10.39DD121 pKa = 4.23SQPSGTLPAAHH132 pKa = 7.37GIFDD136 pKa = 6.05LIDD139 pKa = 3.56SFTDD143 pKa = 4.54YY144 pKa = 10.59PATWKK149 pKa = 10.34INRR152 pKa = 11.84DD153 pKa = 3.07MGHH156 pKa = 6.44RR157 pKa = 11.84FVIKK161 pKa = 10.17RR162 pKa = 11.84RR163 pKa = 11.84WVNWLEE169 pKa = 3.95ADD171 pKa = 4.24GSISTSNYY179 pKa = 9.46SGAPSAPAKK188 pKa = 10.53SSVVFNKK195 pKa = 9.4FVKK198 pKa = 10.13RR199 pKa = 11.84LGVRR203 pKa = 11.84TEE205 pKa = 3.93WKK207 pKa = 8.31NTTTGGIGDD216 pKa = 3.75VSKK219 pKa = 10.55GALYY223 pKa = 10.2IVVARR228 pKa = 11.84QLYY231 pKa = 10.21DD232 pKa = 3.02IDD234 pKa = 3.42VRR236 pKa = 11.84GRR238 pKa = 11.84LRR240 pKa = 11.84VYY242 pKa = 10.06FKK244 pKa = 11.14SVGNQQ249 pKa = 2.8
Molecular weight: 27.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
952 |
94 |
344 |
238.0 |
26.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.357 ± 0.658 | 1.576 ± 0.415 |
4.202 ± 0.233 | 4.412 ± 0.829 |
4.937 ± 0.477 | 7.248 ± 1.685 |
2.206 ± 0.473 | 6.197 ± 0.919 |
5.777 ± 0.427 | 7.143 ± 0.771 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.471 ± 0.126 | 4.937 ± 0.607 |
6.092 ± 0.596 | 3.992 ± 0.785 |
5.987 ± 0.886 | 10.504 ± 1.124 |
6.197 ± 0.967 | 5.252 ± 1.359 |
1.891 ± 0.327 | 4.622 ± 0.401 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
