
Capybara microvirus Cap1_SP_209
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.09
Get precalculated fractions of proteins
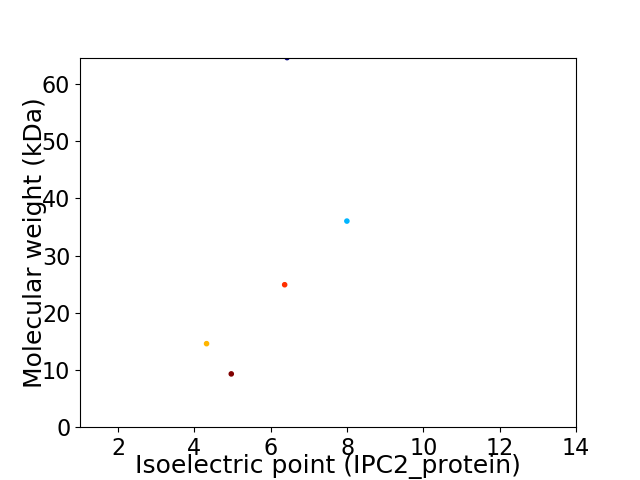
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W852|A0A4P8W852_9VIRU Minor capsid protein OS=Capybara microvirus Cap1_SP_209 OX=2585410 PE=4 SV=1
MM1 pKa = 6.78TAFRR5 pKa = 11.84SRR7 pKa = 11.84FDD9 pKa = 3.58YY10 pKa = 11.54GEE12 pKa = 3.88MPGTVNSEE20 pKa = 3.71PSMTIQSEE28 pKa = 4.07ADD30 pKa = 3.56STSVEE35 pKa = 3.93YY36 pKa = 10.75ALRR39 pKa = 11.84RR40 pKa = 11.84FNATGILGDD49 pKa = 3.88PDD51 pKa = 3.71RR52 pKa = 11.84LASAAYY58 pKa = 10.29LDD60 pKa = 4.04VSDD63 pKa = 4.39VQDD66 pKa = 4.63FEE68 pKa = 4.42QSCNVVARR76 pKa = 11.84ARR78 pKa = 11.84QVFEE82 pKa = 4.36ALPSEE87 pKa = 4.39EE88 pKa = 3.59RR89 pKa = 11.84RR90 pKa = 11.84KK91 pKa = 10.86YY92 pKa = 10.54GDD94 pKa = 3.3SLQVYY99 pKa = 8.28MEE101 pKa = 4.24SRR103 pKa = 11.84LEE105 pKa = 4.2EE106 pKa = 3.89IAQVSGAPSPVVEE119 pKa = 4.61EE120 pKa = 4.75TPSSAAPDD128 pKa = 3.33AAAKK132 pKa = 10.32VDD134 pKa = 3.48
MM1 pKa = 6.78TAFRR5 pKa = 11.84SRR7 pKa = 11.84FDD9 pKa = 3.58YY10 pKa = 11.54GEE12 pKa = 3.88MPGTVNSEE20 pKa = 3.71PSMTIQSEE28 pKa = 4.07ADD30 pKa = 3.56STSVEE35 pKa = 3.93YY36 pKa = 10.75ALRR39 pKa = 11.84RR40 pKa = 11.84FNATGILGDD49 pKa = 3.88PDD51 pKa = 3.71RR52 pKa = 11.84LASAAYY58 pKa = 10.29LDD60 pKa = 4.04VSDD63 pKa = 4.39VQDD66 pKa = 4.63FEE68 pKa = 4.42QSCNVVARR76 pKa = 11.84ARR78 pKa = 11.84QVFEE82 pKa = 4.36ALPSEE87 pKa = 4.39EE88 pKa = 3.59RR89 pKa = 11.84RR90 pKa = 11.84KK91 pKa = 10.86YY92 pKa = 10.54GDD94 pKa = 3.3SLQVYY99 pKa = 8.28MEE101 pKa = 4.24SRR103 pKa = 11.84LEE105 pKa = 4.2EE106 pKa = 3.89IAQVSGAPSPVVEE119 pKa = 4.61EE120 pKa = 4.75TPSSAAPDD128 pKa = 3.33AAAKK132 pKa = 10.32VDD134 pKa = 3.48
Molecular weight: 14.61 kDa
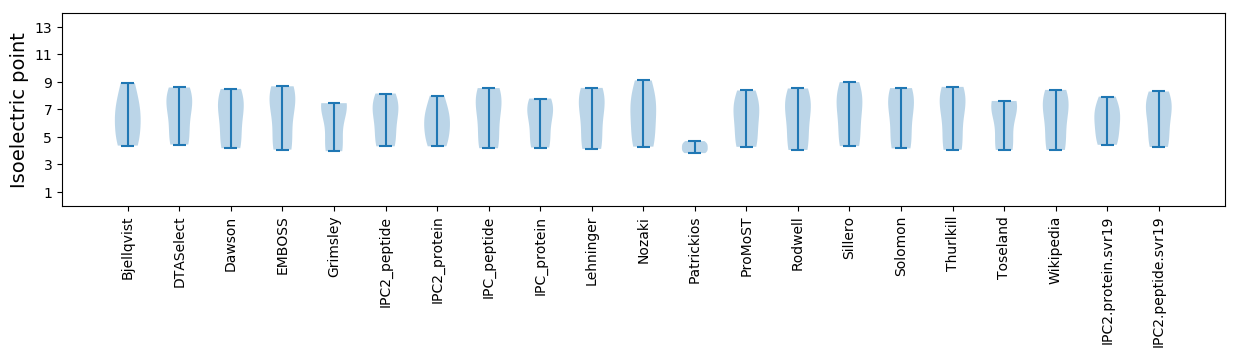
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W5N3|A0A4P8W5N3_9VIRU Minor capsid protein OS=Capybara microvirus Cap1_SP_209 OX=2585410 PE=4 SV=1
MM1 pKa = 7.61PCYY4 pKa = 10.32HH5 pKa = 7.36PVQAWRR11 pKa = 11.84VRR13 pKa = 11.84SGRR16 pKa = 11.84NEE18 pKa = 3.72NGKK21 pKa = 8.87WPITFQFKK29 pKa = 10.02QGYY32 pKa = 7.92ADD34 pKa = 3.46KK35 pKa = 10.24EE36 pKa = 3.96IRR38 pKa = 11.84VPCGQCIGCRR48 pKa = 11.84LEE50 pKa = 4.42KK51 pKa = 10.69SRR53 pKa = 11.84VWALRR58 pKa = 11.84CLHH61 pKa = 6.72EE62 pKa = 4.64SQLHH66 pKa = 4.66EE67 pKa = 4.14QCCFLTLTYY76 pKa = 10.18RR77 pKa = 11.84DD78 pKa = 3.74NPGTLVLRR86 pKa = 11.84DD87 pKa = 3.48LQNFFKK93 pKa = 10.73RR94 pKa = 11.84LRR96 pKa = 11.84KK97 pKa = 9.52AIYY100 pKa = 8.09PIKK103 pKa = 9.97VRR105 pKa = 11.84YY106 pKa = 7.28YY107 pKa = 9.44ACGEE111 pKa = 3.99YY112 pKa = 10.56GSNFGRR118 pKa = 11.84PHH120 pKa = 4.66YY121 pKa = 10.41HH122 pKa = 6.81CLLFGYY128 pKa = 9.97DD129 pKa = 3.72FSRR132 pKa = 11.84DD133 pKa = 2.96RR134 pKa = 11.84VLFKK138 pKa = 10.89RR139 pKa = 11.84SGTNALYY146 pKa = 10.41SSRR149 pKa = 11.84LLDD152 pKa = 3.7SLWTHH157 pKa = 6.86GYY159 pKa = 10.34CLIGDD164 pKa = 3.82VSFDD168 pKa = 3.02SCAYY172 pKa = 7.89VARR175 pKa = 11.84YY176 pKa = 8.43CVKK179 pKa = 10.42KK180 pKa = 9.35VTGEE184 pKa = 4.21GKK186 pKa = 9.7VEE188 pKa = 4.36HH189 pKa = 6.62YY190 pKa = 10.8VDD192 pKa = 4.84PDD194 pKa = 3.29TGEE197 pKa = 3.72LRR199 pKa = 11.84KK200 pKa = 10.0PEE202 pKa = 4.01FAVMSRR208 pKa = 11.84RR209 pKa = 11.84PGIGLDD215 pKa = 2.73WFLRR219 pKa = 11.84YY220 pKa = 9.12RR221 pKa = 11.84DD222 pKa = 3.22EE223 pKa = 4.48VMAHH227 pKa = 6.66EE228 pKa = 4.96GMCYY232 pKa = 9.92INGVFCKK239 pKa = 10.1IPRR242 pKa = 11.84YY243 pKa = 9.69YY244 pKa = 10.35EE245 pKa = 4.14EE246 pKa = 5.47KK247 pKa = 10.46DD248 pKa = 3.11ACAGNDD254 pKa = 3.65GLVKK258 pKa = 10.49LRR260 pKa = 11.84QARR263 pKa = 11.84AAAARR268 pKa = 11.84RR269 pKa = 11.84VQDD272 pKa = 3.59MGEE275 pKa = 3.89QSPDD279 pKa = 2.88RR280 pKa = 11.84LAVRR284 pKa = 11.84EE285 pKa = 4.19EE286 pKa = 4.03CAEE289 pKa = 4.09MNLARR294 pKa = 11.84LKK296 pKa = 10.63RR297 pKa = 11.84GYY299 pKa = 10.61EE300 pKa = 3.85EE301 pKa = 3.96TASFLQPFEE310 pKa = 4.09
MM1 pKa = 7.61PCYY4 pKa = 10.32HH5 pKa = 7.36PVQAWRR11 pKa = 11.84VRR13 pKa = 11.84SGRR16 pKa = 11.84NEE18 pKa = 3.72NGKK21 pKa = 8.87WPITFQFKK29 pKa = 10.02QGYY32 pKa = 7.92ADD34 pKa = 3.46KK35 pKa = 10.24EE36 pKa = 3.96IRR38 pKa = 11.84VPCGQCIGCRR48 pKa = 11.84LEE50 pKa = 4.42KK51 pKa = 10.69SRR53 pKa = 11.84VWALRR58 pKa = 11.84CLHH61 pKa = 6.72EE62 pKa = 4.64SQLHH66 pKa = 4.66EE67 pKa = 4.14QCCFLTLTYY76 pKa = 10.18RR77 pKa = 11.84DD78 pKa = 3.74NPGTLVLRR86 pKa = 11.84DD87 pKa = 3.48LQNFFKK93 pKa = 10.73RR94 pKa = 11.84LRR96 pKa = 11.84KK97 pKa = 9.52AIYY100 pKa = 8.09PIKK103 pKa = 9.97VRR105 pKa = 11.84YY106 pKa = 7.28YY107 pKa = 9.44ACGEE111 pKa = 3.99YY112 pKa = 10.56GSNFGRR118 pKa = 11.84PHH120 pKa = 4.66YY121 pKa = 10.41HH122 pKa = 6.81CLLFGYY128 pKa = 9.97DD129 pKa = 3.72FSRR132 pKa = 11.84DD133 pKa = 2.96RR134 pKa = 11.84VLFKK138 pKa = 10.89RR139 pKa = 11.84SGTNALYY146 pKa = 10.41SSRR149 pKa = 11.84LLDD152 pKa = 3.7SLWTHH157 pKa = 6.86GYY159 pKa = 10.34CLIGDD164 pKa = 3.82VSFDD168 pKa = 3.02SCAYY172 pKa = 7.89VARR175 pKa = 11.84YY176 pKa = 8.43CVKK179 pKa = 10.42KK180 pKa = 9.35VTGEE184 pKa = 4.21GKK186 pKa = 9.7VEE188 pKa = 4.36HH189 pKa = 6.62YY190 pKa = 10.8VDD192 pKa = 4.84PDD194 pKa = 3.29TGEE197 pKa = 3.72LRR199 pKa = 11.84KK200 pKa = 10.0PEE202 pKa = 4.01FAVMSRR208 pKa = 11.84RR209 pKa = 11.84PGIGLDD215 pKa = 2.73WFLRR219 pKa = 11.84YY220 pKa = 9.12RR221 pKa = 11.84DD222 pKa = 3.22EE223 pKa = 4.48VMAHH227 pKa = 6.66EE228 pKa = 4.96GMCYY232 pKa = 9.92INGVFCKK239 pKa = 10.1IPRR242 pKa = 11.84YY243 pKa = 9.69YY244 pKa = 10.35EE245 pKa = 4.14EE246 pKa = 5.47KK247 pKa = 10.46DD248 pKa = 3.11ACAGNDD254 pKa = 3.65GLVKK258 pKa = 10.49LRR260 pKa = 11.84QARR263 pKa = 11.84AAAARR268 pKa = 11.84RR269 pKa = 11.84VQDD272 pKa = 3.59MGEE275 pKa = 3.89QSPDD279 pKa = 2.88RR280 pKa = 11.84LAVRR284 pKa = 11.84EE285 pKa = 4.19EE286 pKa = 4.03CAEE289 pKa = 4.09MNLARR294 pKa = 11.84LKK296 pKa = 10.63RR297 pKa = 11.84GYY299 pKa = 10.61EE300 pKa = 3.85EE301 pKa = 3.96TASFLQPFEE310 pKa = 4.09
Molecular weight: 36.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1341 |
85 |
578 |
268.2 |
29.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.799 ± 1.944 | 1.342 ± 1.041 |
6.115 ± 0.547 | 4.847 ± 1.026 |
4.773 ± 0.446 | 7.383 ± 0.758 |
1.79 ± 0.362 | 3.505 ± 0.845 |
3.579 ± 0.581 | 8.352 ± 0.528 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.237 ± 0.67 | 4.549 ± 0.751 |
4.847 ± 0.819 | 4.773 ± 0.476 |
7.084 ± 0.947 | 8.949 ± 1.317 |
5.444 ± 1.178 | 6.04 ± 0.847 |
1.566 ± 0.332 | 4.027 ± 0.601 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
