
Capybara microvirus Cap3_SP_319
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.81
Get precalculated fractions of proteins
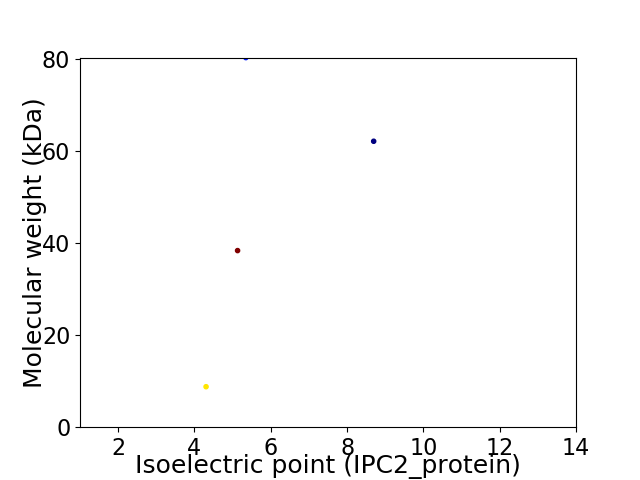
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
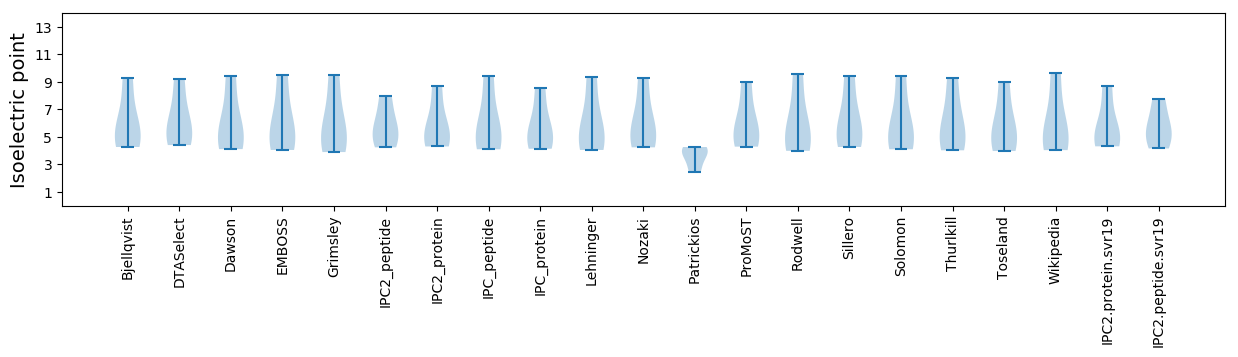
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V1FVS0|A0A4V1FVS0_9VIRU Uncharacterized protein OS=Capybara microvirus Cap3_SP_319 OX=2585427 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 10.47NQGSPILVRR11 pKa = 11.84HH12 pKa = 5.76YY13 pKa = 11.63SDD15 pKa = 3.64FHH17 pKa = 7.56QNFISSPLSEE27 pKa = 4.43FQNSLGSVDD36 pKa = 4.89DD37 pKa = 4.36YY38 pKa = 11.92SFEE41 pKa = 4.29AQLSSGLPLGSVNPTAFLGSPLSEE65 pKa = 3.71EE66 pKa = 4.54DD67 pKa = 3.4ANKK70 pKa = 10.35QITDD74 pKa = 3.14IVEE77 pKa = 4.66SINSS81 pKa = 3.49
MM1 pKa = 7.67KK2 pKa = 10.47NQGSPILVRR11 pKa = 11.84HH12 pKa = 5.76YY13 pKa = 11.63SDD15 pKa = 3.64FHH17 pKa = 7.56QNFISSPLSEE27 pKa = 4.43FQNSLGSVDD36 pKa = 4.89DD37 pKa = 4.36YY38 pKa = 11.92SFEE41 pKa = 4.29AQLSSGLPLGSVNPTAFLGSPLSEE65 pKa = 3.71EE66 pKa = 4.54DD67 pKa = 3.4ANKK70 pKa = 10.35QITDD74 pKa = 3.14IVEE77 pKa = 4.66SINSS81 pKa = 3.49
Molecular weight: 8.8 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W8B3|A0A4P8W8B3_9VIRU Major capsid protein OS=Capybara microvirus Cap3_SP_319 OX=2585427 PE=4 SV=1
MM1 pKa = 7.81PCFTHH6 pKa = 6.14KK7 pKa = 9.67TLFNRR12 pKa = 11.84VSRR15 pKa = 11.84HH16 pKa = 5.27LALTDD21 pKa = 3.62KK22 pKa = 10.63PFLEE26 pKa = 4.91AGCGVCHH33 pKa = 7.41DD34 pKa = 4.66CLSKK38 pKa = 10.85KK39 pKa = 10.29RR40 pKa = 11.84FSLYY44 pKa = 8.92IRR46 pKa = 11.84VWFEE50 pKa = 3.67VEE52 pKa = 5.12DD53 pKa = 3.46ILKK56 pKa = 10.66RR57 pKa = 11.84GGFVLFPTYY66 pKa = 10.25TYY68 pKa = 11.16DD69 pKa = 5.19DD70 pKa = 3.4SHH72 pKa = 8.47LPYY75 pKa = 10.03FDD77 pKa = 4.64SFSHH81 pKa = 5.0QLSYY85 pKa = 11.42GVVDD89 pKa = 3.75SRR91 pKa = 11.84DD92 pKa = 3.52YY93 pKa = 11.35KK94 pKa = 11.16LNEE97 pKa = 4.08VPCFDD102 pKa = 3.21MSVFKK107 pKa = 10.81GSKK110 pKa = 10.32GLVKK114 pKa = 10.56NLRR117 pKa = 11.84NYY119 pKa = 10.69AYY121 pKa = 10.28RR122 pKa = 11.84KK123 pKa = 8.62YY124 pKa = 10.22HH125 pKa = 5.99VDD127 pKa = 4.04GIKK130 pKa = 10.8YY131 pKa = 8.44LVCPEE136 pKa = 4.39YY137 pKa = 11.02GPSDD141 pKa = 3.68GSTHH145 pKa = 6.43RR146 pKa = 11.84PHH148 pKa = 6.12YY149 pKa = 10.3HH150 pKa = 6.81VMFFCPPCEE159 pKa = 4.02GGEE162 pKa = 4.54CVNSVYY168 pKa = 10.67RR169 pKa = 11.84QDD171 pKa = 3.66LTRR174 pKa = 11.84LALSLWQDD182 pKa = 2.84KK183 pKa = 10.71RR184 pKa = 11.84KK185 pKa = 10.41LGFADD190 pKa = 3.71YY191 pKa = 10.74SRR193 pKa = 11.84EE194 pKa = 3.65FGPYY198 pKa = 8.12VTSLQAAKK206 pKa = 10.37YY207 pKa = 8.61ISKK210 pKa = 10.52YY211 pKa = 6.51CTKK214 pKa = 10.53DD215 pKa = 2.79ISEE218 pKa = 4.43WLSVSPQLKK227 pKa = 9.5SYY229 pKa = 10.91LYY231 pKa = 8.62GTSSVIDD238 pKa = 3.83RR239 pKa = 11.84NNLFLIRR246 pKa = 11.84SYY248 pKa = 10.95LPKK251 pKa = 10.21PLWSDD256 pKa = 3.37GLGISMVDD264 pKa = 3.39YY265 pKa = 10.97LKK267 pKa = 11.2SLDD270 pKa = 3.52GDD272 pKa = 4.15SFVSAFTDD280 pKa = 4.62GVHH283 pKa = 5.71VPKK286 pKa = 10.65DD287 pKa = 3.81FKK289 pKa = 11.44EE290 pKa = 4.37DD291 pKa = 3.3GSKK294 pKa = 10.78NFYY297 pKa = 10.47SVPRR301 pKa = 11.84YY302 pKa = 9.56IYY304 pKa = 11.1NKK306 pKa = 9.67VLYY309 pKa = 10.06DD310 pKa = 3.76KK311 pKa = 11.43VGDD314 pKa = 3.77SYY316 pKa = 11.79LPNEE320 pKa = 4.45FNHH323 pKa = 6.83GYY325 pKa = 9.0IKK327 pKa = 10.6RR328 pKa = 11.84IISRR332 pKa = 11.84KK333 pKa = 9.33LDD335 pKa = 3.48LSFEE339 pKa = 4.33RR340 pKa = 11.84FNSDD344 pKa = 3.66LLNASIVSDD353 pKa = 3.21NGKK356 pKa = 10.19NYY358 pKa = 10.51NAFFNNLVSSYY369 pKa = 10.25GKK371 pKa = 10.09EE372 pKa = 3.55YY373 pKa = 11.02LFFSRR378 pKa = 11.84YY379 pKa = 9.36LYY381 pKa = 10.21NVPLSWFSSHH391 pKa = 7.67DD392 pKa = 3.08IDD394 pKa = 5.69VNVLDD399 pKa = 4.28SSSIRR404 pKa = 11.84DD405 pKa = 3.47VVVSYY410 pKa = 11.01LVDD413 pKa = 3.74CSLKK417 pKa = 10.4PRR419 pKa = 11.84FIYY422 pKa = 9.63PSKK425 pKa = 10.17PWYY428 pKa = 10.2KK429 pKa = 10.41SDD431 pKa = 4.18GFILGKK437 pKa = 10.15NIHH440 pKa = 6.14RR441 pKa = 11.84LGYY444 pKa = 9.45NVWVSDD450 pKa = 3.11GKK452 pKa = 10.88IFIRR456 pKa = 11.84SPRR459 pKa = 11.84NPQKK463 pKa = 11.01LIFNRR468 pKa = 11.84DD469 pKa = 3.18SFNSLDD475 pKa = 3.72YY476 pKa = 10.64FRR478 pKa = 11.84PPLIQHH484 pKa = 6.56FFFIWKK490 pKa = 8.83SYY492 pKa = 9.93KK493 pKa = 10.18VALQKK498 pKa = 11.17ASLLKK503 pKa = 10.88SKK505 pKa = 8.77TLQDD509 pKa = 3.22KK510 pKa = 10.75GKK512 pKa = 10.56AFYY515 pKa = 9.52THH517 pKa = 5.97VSTRR521 pKa = 11.84KK522 pKa = 9.29FRR524 pKa = 11.84PLKK527 pKa = 10.43FNSLTFTT534 pKa = 3.89
MM1 pKa = 7.81PCFTHH6 pKa = 6.14KK7 pKa = 9.67TLFNRR12 pKa = 11.84VSRR15 pKa = 11.84HH16 pKa = 5.27LALTDD21 pKa = 3.62KK22 pKa = 10.63PFLEE26 pKa = 4.91AGCGVCHH33 pKa = 7.41DD34 pKa = 4.66CLSKK38 pKa = 10.85KK39 pKa = 10.29RR40 pKa = 11.84FSLYY44 pKa = 8.92IRR46 pKa = 11.84VWFEE50 pKa = 3.67VEE52 pKa = 5.12DD53 pKa = 3.46ILKK56 pKa = 10.66RR57 pKa = 11.84GGFVLFPTYY66 pKa = 10.25TYY68 pKa = 11.16DD69 pKa = 5.19DD70 pKa = 3.4SHH72 pKa = 8.47LPYY75 pKa = 10.03FDD77 pKa = 4.64SFSHH81 pKa = 5.0QLSYY85 pKa = 11.42GVVDD89 pKa = 3.75SRR91 pKa = 11.84DD92 pKa = 3.52YY93 pKa = 11.35KK94 pKa = 11.16LNEE97 pKa = 4.08VPCFDD102 pKa = 3.21MSVFKK107 pKa = 10.81GSKK110 pKa = 10.32GLVKK114 pKa = 10.56NLRR117 pKa = 11.84NYY119 pKa = 10.69AYY121 pKa = 10.28RR122 pKa = 11.84KK123 pKa = 8.62YY124 pKa = 10.22HH125 pKa = 5.99VDD127 pKa = 4.04GIKK130 pKa = 10.8YY131 pKa = 8.44LVCPEE136 pKa = 4.39YY137 pKa = 11.02GPSDD141 pKa = 3.68GSTHH145 pKa = 6.43RR146 pKa = 11.84PHH148 pKa = 6.12YY149 pKa = 10.3HH150 pKa = 6.81VMFFCPPCEE159 pKa = 4.02GGEE162 pKa = 4.54CVNSVYY168 pKa = 10.67RR169 pKa = 11.84QDD171 pKa = 3.66LTRR174 pKa = 11.84LALSLWQDD182 pKa = 2.84KK183 pKa = 10.71RR184 pKa = 11.84KK185 pKa = 10.41LGFADD190 pKa = 3.71YY191 pKa = 10.74SRR193 pKa = 11.84EE194 pKa = 3.65FGPYY198 pKa = 8.12VTSLQAAKK206 pKa = 10.37YY207 pKa = 8.61ISKK210 pKa = 10.52YY211 pKa = 6.51CTKK214 pKa = 10.53DD215 pKa = 2.79ISEE218 pKa = 4.43WLSVSPQLKK227 pKa = 9.5SYY229 pKa = 10.91LYY231 pKa = 8.62GTSSVIDD238 pKa = 3.83RR239 pKa = 11.84NNLFLIRR246 pKa = 11.84SYY248 pKa = 10.95LPKK251 pKa = 10.21PLWSDD256 pKa = 3.37GLGISMVDD264 pKa = 3.39YY265 pKa = 10.97LKK267 pKa = 11.2SLDD270 pKa = 3.52GDD272 pKa = 4.15SFVSAFTDD280 pKa = 4.62GVHH283 pKa = 5.71VPKK286 pKa = 10.65DD287 pKa = 3.81FKK289 pKa = 11.44EE290 pKa = 4.37DD291 pKa = 3.3GSKK294 pKa = 10.78NFYY297 pKa = 10.47SVPRR301 pKa = 11.84YY302 pKa = 9.56IYY304 pKa = 11.1NKK306 pKa = 9.67VLYY309 pKa = 10.06DD310 pKa = 3.76KK311 pKa = 11.43VGDD314 pKa = 3.77SYY316 pKa = 11.79LPNEE320 pKa = 4.45FNHH323 pKa = 6.83GYY325 pKa = 9.0IKK327 pKa = 10.6RR328 pKa = 11.84IISRR332 pKa = 11.84KK333 pKa = 9.33LDD335 pKa = 3.48LSFEE339 pKa = 4.33RR340 pKa = 11.84FNSDD344 pKa = 3.66LLNASIVSDD353 pKa = 3.21NGKK356 pKa = 10.19NYY358 pKa = 10.51NAFFNNLVSSYY369 pKa = 10.25GKK371 pKa = 10.09EE372 pKa = 3.55YY373 pKa = 11.02LFFSRR378 pKa = 11.84YY379 pKa = 9.36LYY381 pKa = 10.21NVPLSWFSSHH391 pKa = 7.67DD392 pKa = 3.08IDD394 pKa = 5.69VNVLDD399 pKa = 4.28SSSIRR404 pKa = 11.84DD405 pKa = 3.47VVVSYY410 pKa = 11.01LVDD413 pKa = 3.74CSLKK417 pKa = 10.4PRR419 pKa = 11.84FIYY422 pKa = 9.63PSKK425 pKa = 10.17PWYY428 pKa = 10.2KK429 pKa = 10.41SDD431 pKa = 4.18GFILGKK437 pKa = 10.15NIHH440 pKa = 6.14RR441 pKa = 11.84LGYY444 pKa = 9.45NVWVSDD450 pKa = 3.11GKK452 pKa = 10.88IFIRR456 pKa = 11.84SPRR459 pKa = 11.84NPQKK463 pKa = 11.01LIFNRR468 pKa = 11.84DD469 pKa = 3.18SFNSLDD475 pKa = 3.72YY476 pKa = 10.64FRR478 pKa = 11.84PPLIQHH484 pKa = 6.56FFFIWKK490 pKa = 8.83SYY492 pKa = 9.93KK493 pKa = 10.18VALQKK498 pKa = 11.17ASLLKK503 pKa = 10.88SKK505 pKa = 8.77TLQDD509 pKa = 3.22KK510 pKa = 10.75GKK512 pKa = 10.56AFYY515 pKa = 9.52THH517 pKa = 5.97VSTRR521 pKa = 11.84KK522 pKa = 9.29FRR524 pKa = 11.84PLKK527 pKa = 10.43FNSLTFTT534 pKa = 3.89
Molecular weight: 62.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1683 |
81 |
710 |
420.8 |
47.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.051 ± 2.056 | 1.307 ± 0.398 |
5.882 ± 0.63 | 4.04 ± 0.766 |
6.595 ± 1.259 | 6.298 ± 0.51 |
2.08 ± 0.556 | 5.169 ± 0.358 |
5.882 ± 0.941 | 9.269 ± 0.506 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.426 ± 0.59 | 7.605 ± 1.184 |
4.159 ± 0.651 | 3.09 ± 1.046 |
3.624 ± 0.77 | 11.23 ± 0.528 |
4.219 ± 1.02 | 6.358 ± 0.353 |
1.01 ± 0.214 | 5.704 ± 1.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
