
Tetraselmis viridis virus S1
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified dsDNA viruses
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
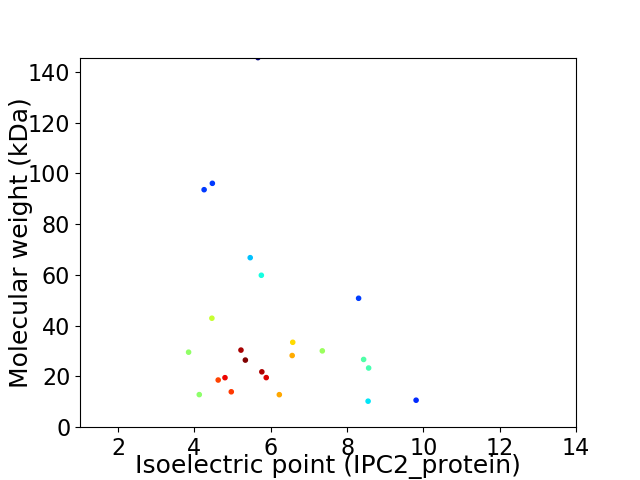
Virtual 2D-PAGE plot for 24 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M4QR70|M4QR70_9VIRU Uncharacterized protein OS=Tetraselmis viridis virus S1 OX=756285 GN=TVSG_00009 PE=4 SV=1
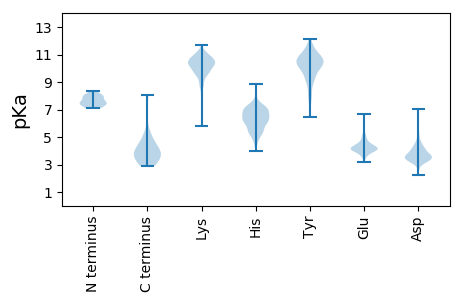
MM1 pKa = 7.16ATIGTRR7 pKa = 11.84AFLATDD13 pKa = 4.31GNWQRR18 pKa = 11.84QYY20 pKa = 11.7DD21 pKa = 4.03NNGNRR26 pKa = 11.84IDD28 pKa = 4.47SIWVPGNNPRR38 pKa = 11.84EE39 pKa = 4.12VNNRR43 pKa = 11.84YY44 pKa = 7.71LTRR47 pKa = 11.84DD48 pKa = 2.99RR49 pKa = 11.84NMRR52 pKa = 11.84AVGAAILPRR61 pKa = 11.84EE62 pKa = 4.14IQDD65 pKa = 4.05KK66 pKa = 10.58IEE68 pKa = 3.97MMALEE73 pKa = 4.93LKK75 pKa = 10.37SDD77 pKa = 3.36QLTAEE82 pKa = 4.07RR83 pKa = 11.84LEE85 pKa = 4.09YY86 pKa = 11.05LNDD89 pKa = 3.11EE90 pKa = 4.22ATPVNEE96 pKa = 4.24EE97 pKa = 3.91VEE99 pKa = 4.3EE100 pKa = 4.31LSDD103 pKa = 3.63YY104 pKa = 11.5DD105 pKa = 3.64KK106 pKa = 11.52DD107 pKa = 3.92VIYY110 pKa = 10.51GHH112 pKa = 6.33VYY114 pKa = 10.86EE115 pKa = 6.09NFDD118 pKa = 3.3LWFSEE123 pKa = 4.16PEE125 pKa = 4.0YY126 pKa = 10.41PEE128 pKa = 3.89WSPTLIEE135 pKa = 4.78FLRR138 pKa = 11.84QNDD141 pKa = 3.71FNYY144 pKa = 10.72DD145 pKa = 3.2EE146 pKa = 4.7YY147 pKa = 11.06EE148 pKa = 3.93ITEE151 pKa = 4.25EE152 pKa = 4.29LPSVIDD158 pKa = 4.17GIYY161 pKa = 9.6IGRR164 pKa = 11.84NVIYY168 pKa = 10.36GRR170 pKa = 11.84YY171 pKa = 7.99LHH173 pKa = 6.93KK174 pKa = 10.8FQDD177 pKa = 5.0DD178 pKa = 4.22DD179 pKa = 4.25FCDD182 pKa = 5.28DD183 pKa = 3.85IQNLAANLGVTLHH196 pKa = 5.98TWADD200 pKa = 2.9IDD202 pKa = 4.25SFCKK206 pKa = 9.99NVYY209 pKa = 10.11IPVALILPPILINDD223 pKa = 3.89DD224 pKa = 4.6PNWGMDD230 pKa = 3.63DD231 pKa = 5.12DD232 pKa = 4.51EE233 pKa = 6.4VIDD236 pKa = 5.51LVSDD240 pKa = 3.94DD241 pKa = 4.94DD242 pKa = 3.82ATIIISDD249 pKa = 3.96SDD251 pKa = 4.16DD252 pKa = 3.49EE253 pKa = 4.42PMM255 pKa = 5.24
MM1 pKa = 7.16ATIGTRR7 pKa = 11.84AFLATDD13 pKa = 4.31GNWQRR18 pKa = 11.84QYY20 pKa = 11.7DD21 pKa = 4.03NNGNRR26 pKa = 11.84IDD28 pKa = 4.47SIWVPGNNPRR38 pKa = 11.84EE39 pKa = 4.12VNNRR43 pKa = 11.84YY44 pKa = 7.71LTRR47 pKa = 11.84DD48 pKa = 2.99RR49 pKa = 11.84NMRR52 pKa = 11.84AVGAAILPRR61 pKa = 11.84EE62 pKa = 4.14IQDD65 pKa = 4.05KK66 pKa = 10.58IEE68 pKa = 3.97MMALEE73 pKa = 4.93LKK75 pKa = 10.37SDD77 pKa = 3.36QLTAEE82 pKa = 4.07RR83 pKa = 11.84LEE85 pKa = 4.09YY86 pKa = 11.05LNDD89 pKa = 3.11EE90 pKa = 4.22ATPVNEE96 pKa = 4.24EE97 pKa = 3.91VEE99 pKa = 4.3EE100 pKa = 4.31LSDD103 pKa = 3.63YY104 pKa = 11.5DD105 pKa = 3.64KK106 pKa = 11.52DD107 pKa = 3.92VIYY110 pKa = 10.51GHH112 pKa = 6.33VYY114 pKa = 10.86EE115 pKa = 6.09NFDD118 pKa = 3.3LWFSEE123 pKa = 4.16PEE125 pKa = 4.0YY126 pKa = 10.41PEE128 pKa = 3.89WSPTLIEE135 pKa = 4.78FLRR138 pKa = 11.84QNDD141 pKa = 3.71FNYY144 pKa = 10.72DD145 pKa = 3.2EE146 pKa = 4.7YY147 pKa = 11.06EE148 pKa = 3.93ITEE151 pKa = 4.25EE152 pKa = 4.29LPSVIDD158 pKa = 4.17GIYY161 pKa = 9.6IGRR164 pKa = 11.84NVIYY168 pKa = 10.36GRR170 pKa = 11.84YY171 pKa = 7.99LHH173 pKa = 6.93KK174 pKa = 10.8FQDD177 pKa = 5.0DD178 pKa = 4.22DD179 pKa = 4.25FCDD182 pKa = 5.28DD183 pKa = 3.85IQNLAANLGVTLHH196 pKa = 5.98TWADD200 pKa = 2.9IDD202 pKa = 4.25SFCKK206 pKa = 9.99NVYY209 pKa = 10.11IPVALILPPILINDD223 pKa = 3.89DD224 pKa = 4.6PNWGMDD230 pKa = 3.63DD231 pKa = 5.12DD232 pKa = 4.51EE233 pKa = 6.4VIDD236 pKa = 5.51LVSDD240 pKa = 3.94DD241 pKa = 4.94DD242 pKa = 3.82ATIIISDD249 pKa = 3.96SDD251 pKa = 4.16DD252 pKa = 3.49EE253 pKa = 4.42PMM255 pKa = 5.24
Molecular weight: 29.56 kDa
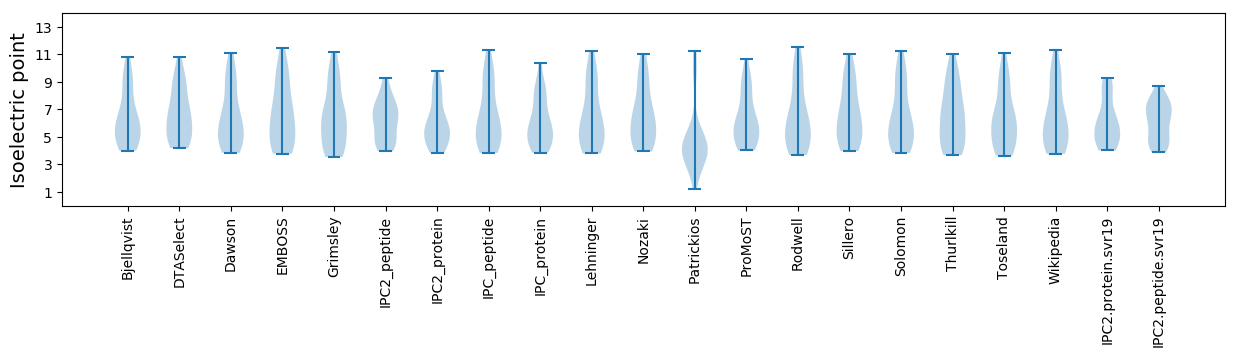
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M4QR85|M4QR85_9VIRU Uncharacterized protein OS=Tetraselmis viridis virus S1 OX=756285 GN=TVSG_00024 PE=4 SV=1
MM1 pKa = 7.79IIGAKK6 pKa = 9.85SGAHH10 pKa = 6.04SGLSAVIGSKK20 pKa = 10.63NNNSHH25 pKa = 4.97MTMGSKK31 pKa = 10.49GGAPSHH37 pKa = 6.77HH38 pKa = 6.07GRR40 pKa = 11.84AVYY43 pKa = 8.49TQANRR48 pKa = 11.84AMGDD52 pKa = 3.17MGGGKK57 pKa = 9.16SAPGNTHH64 pKa = 6.25NSVGGGQRR72 pKa = 11.84DD73 pKa = 3.61APSRR77 pKa = 11.84PDD79 pKa = 4.18GIRR82 pKa = 11.84SMADD86 pKa = 2.92PRR88 pKa = 11.84PSKK91 pKa = 10.14RR92 pKa = 11.84AKK94 pKa = 7.47TTEE97 pKa = 3.81SGKK100 pKa = 10.45KK101 pKa = 9.82NKK103 pKa = 10.38AEE105 pKa = 3.88
MM1 pKa = 7.79IIGAKK6 pKa = 9.85SGAHH10 pKa = 6.04SGLSAVIGSKK20 pKa = 10.63NNNSHH25 pKa = 4.97MTMGSKK31 pKa = 10.49GGAPSHH37 pKa = 6.77HH38 pKa = 6.07GRR40 pKa = 11.84AVYY43 pKa = 8.49TQANRR48 pKa = 11.84AMGDD52 pKa = 3.17MGGGKK57 pKa = 9.16SAPGNTHH64 pKa = 6.25NSVGGGQRR72 pKa = 11.84DD73 pKa = 3.61APSRR77 pKa = 11.84PDD79 pKa = 4.18GIRR82 pKa = 11.84SMADD86 pKa = 2.92PRR88 pKa = 11.84PSKK91 pKa = 10.14RR92 pKa = 11.84AKK94 pKa = 7.47TTEE97 pKa = 3.81SGKK100 pKa = 10.45KK101 pKa = 9.82NKK103 pKa = 10.38AEE105 pKa = 3.88
Molecular weight: 10.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
8262 |
88 |
1274 |
344.3 |
38.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.826 ± 0.421 | 1.041 ± 0.264 |
7.649 ± 0.474 | 6.258 ± 0.381 |
3.546 ± 0.258 | 7.25 ± 0.497 |
2.481 ± 0.203 | 5.507 ± 0.283 |
4.999 ± 0.568 | 6.705 ± 0.371 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.784 ± 0.331 | 5.277 ± 0.386 |
5.507 ± 0.407 | 3.002 ± 0.251 |
5.894 ± 0.51 | 7.286 ± 0.418 |
6.536 ± 0.575 | 6.245 ± 0.395 |
1.198 ± 0.167 | 4.006 ± 0.348 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
