
Lisianthus necrosis virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Procedovirinae; Tombusvirus; Eggplant mottled crinkle virus
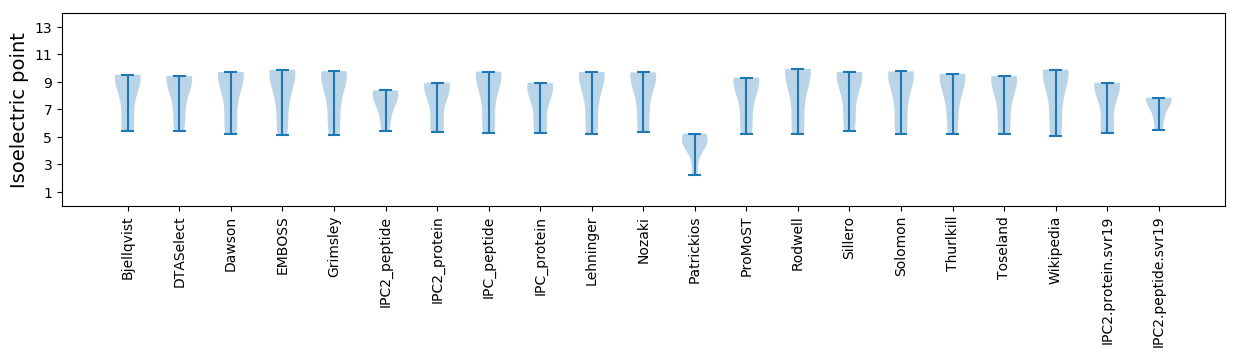
Average proteome isoelectric point is 7.67
Get precalculated fractions of proteins
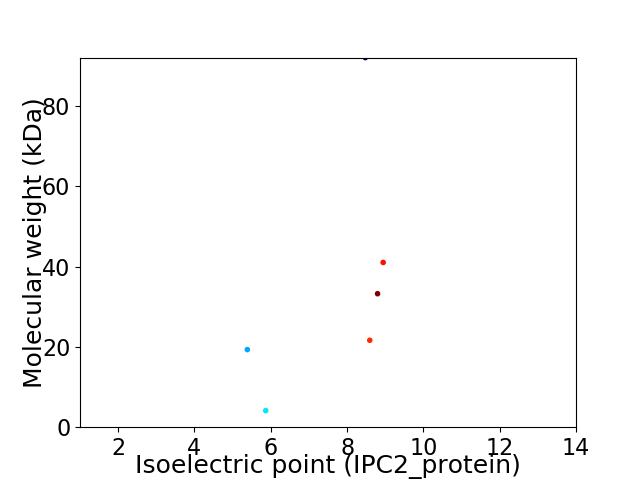
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A7M8Y5|A7M8Y5_9TOMB Uncharacterized protein OS=Lisianthus necrosis virus OX=334425 PE=4 SV=1
MM1 pKa = 7.32EE2 pKa = 5.14RR3 pKa = 11.84AIQGSDD9 pKa = 3.0ARR11 pKa = 11.84EE12 pKa = 3.66QAYY15 pKa = 9.45SEE17 pKa = 4.32RR18 pKa = 11.84WDD20 pKa = 3.91GGCGGTITPFKK31 pKa = 10.79LPDD34 pKa = 3.68EE35 pKa = 4.67SPSLIEE41 pKa = 3.82WRR43 pKa = 11.84LHH45 pKa = 5.17NSEE48 pKa = 4.22EE49 pKa = 5.29SEE51 pKa = 5.27DD52 pKa = 3.69KK53 pKa = 11.15DD54 pKa = 3.57NPLGFKK60 pKa = 10.18EE61 pKa = 4.13SWSFGKK67 pKa = 10.55VVFKK71 pKa = 10.76RR72 pKa = 11.84YY73 pKa = 9.63LRR75 pKa = 11.84YY76 pKa = 10.51DD77 pKa = 3.22GTEE80 pKa = 3.86ASLHH84 pKa = 5.89RR85 pKa = 11.84ALGSWEE91 pKa = 4.03RR92 pKa = 11.84DD93 pKa = 3.57TVNDD97 pKa = 3.45AASRR101 pKa = 11.84FLGFGQIGCTYY112 pKa = 9.88CIRR115 pKa = 11.84FRR117 pKa = 11.84GSCLTISGGSRR128 pKa = 11.84TLQRR132 pKa = 11.84LIEE135 pKa = 3.89MAIRR139 pKa = 11.84TKK141 pKa = 10.37RR142 pKa = 11.84TMLQLAPCEE151 pKa = 4.22VEE153 pKa = 4.75GNVSRR158 pKa = 11.84GSPEE162 pKa = 3.35GTEE165 pKa = 3.95AFKK168 pKa = 11.2EE169 pKa = 4.15EE170 pKa = 4.46SEE172 pKa = 4.25
MM1 pKa = 7.32EE2 pKa = 5.14RR3 pKa = 11.84AIQGSDD9 pKa = 3.0ARR11 pKa = 11.84EE12 pKa = 3.66QAYY15 pKa = 9.45SEE17 pKa = 4.32RR18 pKa = 11.84WDD20 pKa = 3.91GGCGGTITPFKK31 pKa = 10.79LPDD34 pKa = 3.68EE35 pKa = 4.67SPSLIEE41 pKa = 3.82WRR43 pKa = 11.84LHH45 pKa = 5.17NSEE48 pKa = 4.22EE49 pKa = 5.29SEE51 pKa = 5.27DD52 pKa = 3.69KK53 pKa = 11.15DD54 pKa = 3.57NPLGFKK60 pKa = 10.18EE61 pKa = 4.13SWSFGKK67 pKa = 10.55VVFKK71 pKa = 10.76RR72 pKa = 11.84YY73 pKa = 9.63LRR75 pKa = 11.84YY76 pKa = 10.51DD77 pKa = 3.22GTEE80 pKa = 3.86ASLHH84 pKa = 5.89RR85 pKa = 11.84ALGSWEE91 pKa = 4.03RR92 pKa = 11.84DD93 pKa = 3.57TVNDD97 pKa = 3.45AASRR101 pKa = 11.84FLGFGQIGCTYY112 pKa = 9.88CIRR115 pKa = 11.84FRR117 pKa = 11.84GSCLTISGGSRR128 pKa = 11.84TLQRR132 pKa = 11.84LIEE135 pKa = 3.89MAIRR139 pKa = 11.84TKK141 pKa = 10.37RR142 pKa = 11.84TMLQLAPCEE151 pKa = 4.22VEE153 pKa = 4.75GNVSRR158 pKa = 11.84GSPEE162 pKa = 3.35GTEE165 pKa = 3.95AFKK168 pKa = 11.2EE169 pKa = 4.15EE170 pKa = 4.46SEE172 pKa = 4.25
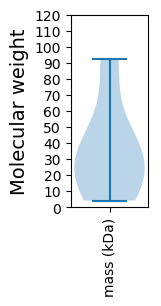
Molecular weight: 19.33 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A7M8Y1|A7M8Y1_9TOMB Isoform of A7M8Y0 RNA-directed RNA polymerase (Fragment) OS=Lisianthus necrosis virus OX=334425 GN=replicase PE=4 SV=1
MM1 pKa = 8.13DD2 pKa = 5.53RR3 pKa = 11.84LLNFLRR9 pKa = 11.84PKK11 pKa = 10.64KK12 pKa = 10.19EE13 pKa = 3.6IFVGDD18 pKa = 3.45FAIGVDD24 pKa = 4.11RR25 pKa = 11.84RR26 pKa = 11.84PSMDD30 pKa = 3.08MFQLVCRR37 pKa = 11.84LALRR41 pKa = 11.84YY42 pKa = 8.41MRR44 pKa = 11.84TGKK47 pKa = 9.76IEE49 pKa = 4.44CNVDD53 pKa = 3.29SLSQFVVEE61 pKa = 5.75LLKK64 pKa = 10.18TDD66 pKa = 5.11CAAKK70 pKa = 10.03WEE72 pKa = 4.24WFMKK76 pKa = 10.02RR77 pKa = 11.84RR78 pKa = 11.84SAGDD82 pKa = 3.25YY83 pKa = 10.33AIPLALAALPIAPLLGYY100 pKa = 7.82ATKK103 pKa = 10.64VRR105 pKa = 11.84TVSVKK110 pKa = 10.79AFGNEE115 pKa = 3.4LTFPFRR121 pKa = 11.84TLRR124 pKa = 11.84PSFPRR129 pKa = 11.84KK130 pKa = 9.85GLLLRR135 pKa = 11.84LAAGLALAPVCALAVYY151 pKa = 7.97ATLPRR156 pKa = 11.84EE157 pKa = 3.94KK158 pKa = 10.69LSVFRR163 pKa = 11.84LRR165 pKa = 11.84TEE167 pKa = 3.98ARR169 pKa = 11.84THH171 pKa = 5.86MEE173 pKa = 4.19DD174 pKa = 3.06EE175 pKa = 4.51RR176 pKa = 11.84EE177 pKa = 4.18ATEE180 pKa = 4.15CLVVEE185 pKa = 4.99PARR188 pKa = 11.84EE189 pKa = 3.99LKK191 pKa = 10.99GKK193 pKa = 10.26DD194 pKa = 3.6GEE196 pKa = 4.5DD197 pKa = 3.55LLTGSRR203 pKa = 11.84MTKK206 pKa = 10.3VIASTGRR213 pKa = 11.84PRR215 pKa = 11.84RR216 pKa = 11.84RR217 pKa = 11.84PYY219 pKa = 9.5AAKK222 pKa = 9.19VAQVARR228 pKa = 11.84AKK230 pKa = 10.53VGYY233 pKa = 9.9LKK235 pKa = 10.75NSPEE239 pKa = 3.62NRR241 pKa = 11.84LIYY244 pKa = 10.08QRR246 pKa = 11.84VIIEE250 pKa = 4.93IMDD253 pKa = 4.1KK254 pKa = 11.13DD255 pKa = 3.69CVRR258 pKa = 11.84YY259 pKa = 9.97VDD261 pKa = 4.33RR262 pKa = 11.84DD263 pKa = 3.8VILPLAIGCCFVYY276 pKa = 10.38PDD278 pKa = 4.17GVEE281 pKa = 3.91EE282 pKa = 5.03SAALWGSSEE291 pKa = 4.13SLGVKK296 pKa = 9.68
MM1 pKa = 8.13DD2 pKa = 5.53RR3 pKa = 11.84LLNFLRR9 pKa = 11.84PKK11 pKa = 10.64KK12 pKa = 10.19EE13 pKa = 3.6IFVGDD18 pKa = 3.45FAIGVDD24 pKa = 4.11RR25 pKa = 11.84RR26 pKa = 11.84PSMDD30 pKa = 3.08MFQLVCRR37 pKa = 11.84LALRR41 pKa = 11.84YY42 pKa = 8.41MRR44 pKa = 11.84TGKK47 pKa = 9.76IEE49 pKa = 4.44CNVDD53 pKa = 3.29SLSQFVVEE61 pKa = 5.75LLKK64 pKa = 10.18TDD66 pKa = 5.11CAAKK70 pKa = 10.03WEE72 pKa = 4.24WFMKK76 pKa = 10.02RR77 pKa = 11.84RR78 pKa = 11.84SAGDD82 pKa = 3.25YY83 pKa = 10.33AIPLALAALPIAPLLGYY100 pKa = 7.82ATKK103 pKa = 10.64VRR105 pKa = 11.84TVSVKK110 pKa = 10.79AFGNEE115 pKa = 3.4LTFPFRR121 pKa = 11.84TLRR124 pKa = 11.84PSFPRR129 pKa = 11.84KK130 pKa = 9.85GLLLRR135 pKa = 11.84LAAGLALAPVCALAVYY151 pKa = 7.97ATLPRR156 pKa = 11.84EE157 pKa = 3.94KK158 pKa = 10.69LSVFRR163 pKa = 11.84LRR165 pKa = 11.84TEE167 pKa = 3.98ARR169 pKa = 11.84THH171 pKa = 5.86MEE173 pKa = 4.19DD174 pKa = 3.06EE175 pKa = 4.51RR176 pKa = 11.84EE177 pKa = 4.18ATEE180 pKa = 4.15CLVVEE185 pKa = 4.99PARR188 pKa = 11.84EE189 pKa = 3.99LKK191 pKa = 10.99GKK193 pKa = 10.26DD194 pKa = 3.6GEE196 pKa = 4.5DD197 pKa = 3.55LLTGSRR203 pKa = 11.84MTKK206 pKa = 10.3VIASTGRR213 pKa = 11.84PRR215 pKa = 11.84RR216 pKa = 11.84RR217 pKa = 11.84PYY219 pKa = 9.5AAKK222 pKa = 9.19VAQVARR228 pKa = 11.84AKK230 pKa = 10.53VGYY233 pKa = 9.9LKK235 pKa = 10.75NSPEE239 pKa = 3.62NRR241 pKa = 11.84LIYY244 pKa = 10.08QRR246 pKa = 11.84VIIEE250 pKa = 4.93IMDD253 pKa = 4.1KK254 pKa = 11.13DD255 pKa = 3.69CVRR258 pKa = 11.84YY259 pKa = 9.97VDD261 pKa = 4.33RR262 pKa = 11.84DD263 pKa = 3.8VILPLAIGCCFVYY276 pKa = 10.38PDD278 pKa = 4.17GVEE281 pKa = 3.91EE282 pKa = 5.03SAALWGSSEE291 pKa = 4.13SLGVKK296 pKa = 9.68
Molecular weight: 33.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
1903 |
39 |
817 |
317.2 |
35.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.04 ± 0.858 | 2.207 ± 0.339 |
4.467 ± 0.299 | 6.043 ± 0.932 |
3.941 ± 0.206 | 7.935 ± 0.662 |
1.576 ± 0.453 | 3.941 ± 0.086 |
5.465 ± 0.643 | 10.194 ± 0.676 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.522 ± 0.127 | 3.573 ± 0.966 |
4.782 ± 0.255 | 2.522 ± 0.239 |
7.725 ± 0.79 | 6.463 ± 0.84 |
6.148 ± 1.158 | 8.04 ± 0.663 |
1.261 ± 0.172 | 3.153 ± 0.204 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
