
Diploscapter pachys
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Nematoda; Chromadorea; Rhabditida; Rhabditina; Rhabditomorpha; Rhabditoidea; Rhabditidae; Rhabditidae incertae sedis; Diploscapter
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
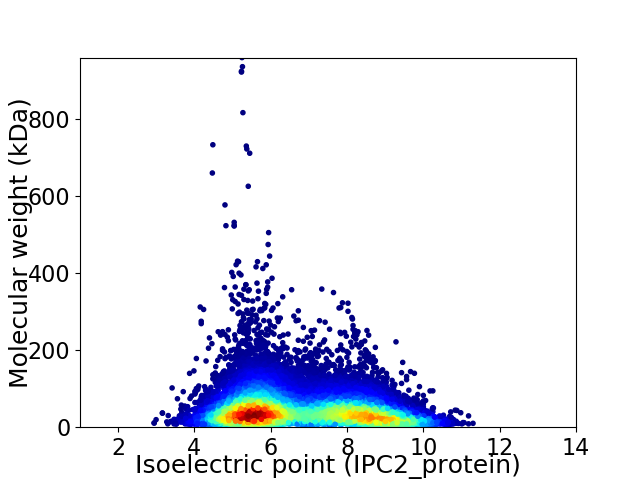
Virtual 2D-PAGE plot for 33981 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2A2KTF8|A0A2A2KTF8_9BILA Uncharacterized protein OS=Diploscapter pachys OX=2018661 GN=WR25_17893 PE=3 SV=1
MM1 pKa = 7.81RR2 pKa = 11.84GGAGADD8 pKa = 3.56RR9 pKa = 11.84FIGGAGNDD17 pKa = 3.64TVSYY21 pKa = 9.22TDD23 pKa = 3.38AGAGITLNFKK33 pKa = 9.82TGVHH37 pKa = 5.77TGIAAGDD44 pKa = 3.73TFDD47 pKa = 5.87SIEE50 pKa = 4.24LFQGSNYY57 pKa = 10.35ADD59 pKa = 3.5TFISDD64 pKa = 3.31ARR66 pKa = 11.84AHH68 pKa = 5.34TFEE71 pKa = 5.95GGLNDD76 pKa = 3.47TMDD79 pKa = 3.66YY80 pKa = 10.8SLSEE84 pKa = 3.9QAVTITIDD92 pKa = 3.26NTGGKK97 pKa = 9.07GQGGDD102 pKa = 3.41AEE104 pKa = 4.54GDD106 pKa = 3.67KK107 pKa = 10.15FTGMYY112 pKa = 10.09SVIGSSQNDD121 pKa = 3.9TFIMQATGSSKK132 pKa = 10.81SFAGGAGDD140 pKa = 4.38DD141 pKa = 3.67VYY143 pKa = 11.62YY144 pKa = 10.78LHH146 pKa = 6.44NTAGVFEE153 pKa = 4.22QAGGGIDD160 pKa = 3.59EE161 pKa = 4.62VRR163 pKa = 11.84TTLASYY169 pKa = 9.36TLTPEE174 pKa = 4.39TGVHH178 pKa = 5.44TGIAAGDD185 pKa = 3.72TFDD188 pKa = 5.35SIEE191 pKa = 4.01VFQGSNYY198 pKa = 10.33ADD200 pKa = 3.5TFISDD205 pKa = 3.31ARR207 pKa = 11.84AHH209 pKa = 5.34TFEE212 pKa = 5.97GGLNDD217 pKa = 3.68TLDD220 pKa = 3.74YY221 pKa = 11.02SLSEE225 pKa = 3.94QAVKK229 pKa = 9.06ITIEE233 pKa = 4.2GNGGSGQGGDD243 pKa = 3.47AEE245 pKa = 4.53GDD247 pKa = 3.67KK248 pKa = 10.15FTGMYY253 pKa = 10.09SVIGSSQNDD262 pKa = 3.9TFIMQATGSSKK273 pKa = 10.71NFAGGAGDD281 pKa = 3.88DD282 pKa = 3.69VYY284 pKa = 11.74YY285 pKa = 10.59LYY287 pKa = 10.05STAGVFEE294 pKa = 4.15QAGGGIDD301 pKa = 3.59EE302 pKa = 4.89VRR304 pKa = 11.84TTLANYY310 pKa = 7.48TLTNEE315 pKa = 4.23VEE317 pKa = 4.09NLTFIGTGNFTGYY330 pKa = 11.14GNASNNVIVGGAGNDD345 pKa = 3.73TLFGGAGADD354 pKa = 3.4RR355 pKa = 11.84FVGGAGIDD363 pKa = 3.58TVSYY367 pKa = 10.53GDD369 pKa = 3.64SSSGILLNLKK379 pKa = 8.67TGQHH383 pKa = 5.57GGIAEE388 pKa = 4.42GDD390 pKa = 3.6TFEE393 pKa = 6.25GIEE396 pKa = 4.18KK397 pKa = 10.4FVGSSLGDD405 pKa = 3.09VFVANDD411 pKa = 3.24QANDD415 pKa = 3.38FNGAGGVDD423 pKa = 4.13LLTFAGEE430 pKa = 4.43GAGITLDD437 pKa = 4.7LNATGNDD444 pKa = 3.45LFTSIEE450 pKa = 4.29SFEE453 pKa = 3.97GTAFNDD459 pKa = 3.89TFIGTAASEE468 pKa = 4.13NFIGGAGADD477 pKa = 4.15VINGGAGRR485 pKa = 11.84DD486 pKa = 3.8SAWYY490 pKa = 8.7ISSNASVSVDD500 pKa = 3.3LATGVVGGGHH510 pKa = 7.01AEE512 pKa = 3.88GDD514 pKa = 3.82VLSGIEE520 pKa = 4.24GVAGSAFADD529 pKa = 3.95VLNGDD534 pKa = 3.82AGANTLYY541 pKa = 10.67GAEE544 pKa = 4.38GDD546 pKa = 4.08DD547 pKa = 4.66VINGGAGNDD556 pKa = 3.22ILYY559 pKa = 10.9GDD561 pKa = 5.43GIYY564 pKa = 9.49STDD567 pKa = 3.47FGALARR573 pKa = 11.84PANTQVAQADD583 pKa = 4.49TINGGDD589 pKa = 3.95GDD591 pKa = 4.16DD592 pKa = 4.65TITGALNDD600 pKa = 4.0TGSIYY605 pKa = 10.83HH606 pKa = 7.02GDD608 pKa = 3.66AGNDD612 pKa = 3.73TIQTANGKK620 pKa = 10.3AYY622 pKa = 10.39GDD624 pKa = 4.37DD625 pKa = 3.93GNDD628 pKa = 2.89RR629 pKa = 11.84LVGNGYY635 pKa = 10.65AYY637 pKa = 10.13EE638 pKa = 4.32LYY640 pKa = 10.67GGAGADD646 pKa = 3.66RR647 pKa = 11.84LDD649 pKa = 3.77MMSAGYY655 pKa = 10.85AFGGEE660 pKa = 4.1GSDD663 pKa = 3.88IYY665 pKa = 10.77TIYY668 pKa = 10.91SKK670 pKa = 11.13AGVVIQDD677 pKa = 3.69TGTAGIDD684 pKa = 3.41VVVLKK689 pKa = 10.82NIQTFQDD696 pKa = 3.36VVVQNDD702 pKa = 3.73GVNAYY707 pKa = 9.3IFSKK711 pKa = 10.97ADD713 pKa = 3.2WNAGNLDD720 pKa = 3.96SGVMLSNWYY729 pKa = 10.08AGSNTIEE736 pKa = 4.08SFQTNNGDD744 pKa = 3.52TFTITT749 pKa = 3.44
MM1 pKa = 7.81RR2 pKa = 11.84GGAGADD8 pKa = 3.56RR9 pKa = 11.84FIGGAGNDD17 pKa = 3.64TVSYY21 pKa = 9.22TDD23 pKa = 3.38AGAGITLNFKK33 pKa = 9.82TGVHH37 pKa = 5.77TGIAAGDD44 pKa = 3.73TFDD47 pKa = 5.87SIEE50 pKa = 4.24LFQGSNYY57 pKa = 10.35ADD59 pKa = 3.5TFISDD64 pKa = 3.31ARR66 pKa = 11.84AHH68 pKa = 5.34TFEE71 pKa = 5.95GGLNDD76 pKa = 3.47TMDD79 pKa = 3.66YY80 pKa = 10.8SLSEE84 pKa = 3.9QAVTITIDD92 pKa = 3.26NTGGKK97 pKa = 9.07GQGGDD102 pKa = 3.41AEE104 pKa = 4.54GDD106 pKa = 3.67KK107 pKa = 10.15FTGMYY112 pKa = 10.09SVIGSSQNDD121 pKa = 3.9TFIMQATGSSKK132 pKa = 10.81SFAGGAGDD140 pKa = 4.38DD141 pKa = 3.67VYY143 pKa = 11.62YY144 pKa = 10.78LHH146 pKa = 6.44NTAGVFEE153 pKa = 4.22QAGGGIDD160 pKa = 3.59EE161 pKa = 4.62VRR163 pKa = 11.84TTLASYY169 pKa = 9.36TLTPEE174 pKa = 4.39TGVHH178 pKa = 5.44TGIAAGDD185 pKa = 3.72TFDD188 pKa = 5.35SIEE191 pKa = 4.01VFQGSNYY198 pKa = 10.33ADD200 pKa = 3.5TFISDD205 pKa = 3.31ARR207 pKa = 11.84AHH209 pKa = 5.34TFEE212 pKa = 5.97GGLNDD217 pKa = 3.68TLDD220 pKa = 3.74YY221 pKa = 11.02SLSEE225 pKa = 3.94QAVKK229 pKa = 9.06ITIEE233 pKa = 4.2GNGGSGQGGDD243 pKa = 3.47AEE245 pKa = 4.53GDD247 pKa = 3.67KK248 pKa = 10.15FTGMYY253 pKa = 10.09SVIGSSQNDD262 pKa = 3.9TFIMQATGSSKK273 pKa = 10.71NFAGGAGDD281 pKa = 3.88DD282 pKa = 3.69VYY284 pKa = 11.74YY285 pKa = 10.59LYY287 pKa = 10.05STAGVFEE294 pKa = 4.15QAGGGIDD301 pKa = 3.59EE302 pKa = 4.89VRR304 pKa = 11.84TTLANYY310 pKa = 7.48TLTNEE315 pKa = 4.23VEE317 pKa = 4.09NLTFIGTGNFTGYY330 pKa = 11.14GNASNNVIVGGAGNDD345 pKa = 3.73TLFGGAGADD354 pKa = 3.4RR355 pKa = 11.84FVGGAGIDD363 pKa = 3.58TVSYY367 pKa = 10.53GDD369 pKa = 3.64SSSGILLNLKK379 pKa = 8.67TGQHH383 pKa = 5.57GGIAEE388 pKa = 4.42GDD390 pKa = 3.6TFEE393 pKa = 6.25GIEE396 pKa = 4.18KK397 pKa = 10.4FVGSSLGDD405 pKa = 3.09VFVANDD411 pKa = 3.24QANDD415 pKa = 3.38FNGAGGVDD423 pKa = 4.13LLTFAGEE430 pKa = 4.43GAGITLDD437 pKa = 4.7LNATGNDD444 pKa = 3.45LFTSIEE450 pKa = 4.29SFEE453 pKa = 3.97GTAFNDD459 pKa = 3.89TFIGTAASEE468 pKa = 4.13NFIGGAGADD477 pKa = 4.15VINGGAGRR485 pKa = 11.84DD486 pKa = 3.8SAWYY490 pKa = 8.7ISSNASVSVDD500 pKa = 3.3LATGVVGGGHH510 pKa = 7.01AEE512 pKa = 3.88GDD514 pKa = 3.82VLSGIEE520 pKa = 4.24GVAGSAFADD529 pKa = 3.95VLNGDD534 pKa = 3.82AGANTLYY541 pKa = 10.67GAEE544 pKa = 4.38GDD546 pKa = 4.08DD547 pKa = 4.66VINGGAGNDD556 pKa = 3.22ILYY559 pKa = 10.9GDD561 pKa = 5.43GIYY564 pKa = 9.49STDD567 pKa = 3.47FGALARR573 pKa = 11.84PANTQVAQADD583 pKa = 4.49TINGGDD589 pKa = 3.95GDD591 pKa = 4.16DD592 pKa = 4.65TITGALNDD600 pKa = 4.0TGSIYY605 pKa = 10.83HH606 pKa = 7.02GDD608 pKa = 3.66AGNDD612 pKa = 3.73TIQTANGKK620 pKa = 10.3AYY622 pKa = 10.39GDD624 pKa = 4.37DD625 pKa = 3.93GNDD628 pKa = 2.89RR629 pKa = 11.84LVGNGYY635 pKa = 10.65AYY637 pKa = 10.13EE638 pKa = 4.32LYY640 pKa = 10.67GGAGADD646 pKa = 3.66RR647 pKa = 11.84LDD649 pKa = 3.77MMSAGYY655 pKa = 10.85AFGGEE660 pKa = 4.1GSDD663 pKa = 3.88IYY665 pKa = 10.77TIYY668 pKa = 10.91SKK670 pKa = 11.13AGVVIQDD677 pKa = 3.69TGTAGIDD684 pKa = 3.41VVVLKK689 pKa = 10.82NIQTFQDD696 pKa = 3.36VVVQNDD702 pKa = 3.73GVNAYY707 pKa = 9.3IFSKK711 pKa = 10.97ADD713 pKa = 3.2WNAGNLDD720 pKa = 3.96SGVMLSNWYY729 pKa = 10.08AGSNTIEE736 pKa = 4.08SFQTNNGDD744 pKa = 3.52TFTITT749 pKa = 3.44
Molecular weight: 76.05 kDa
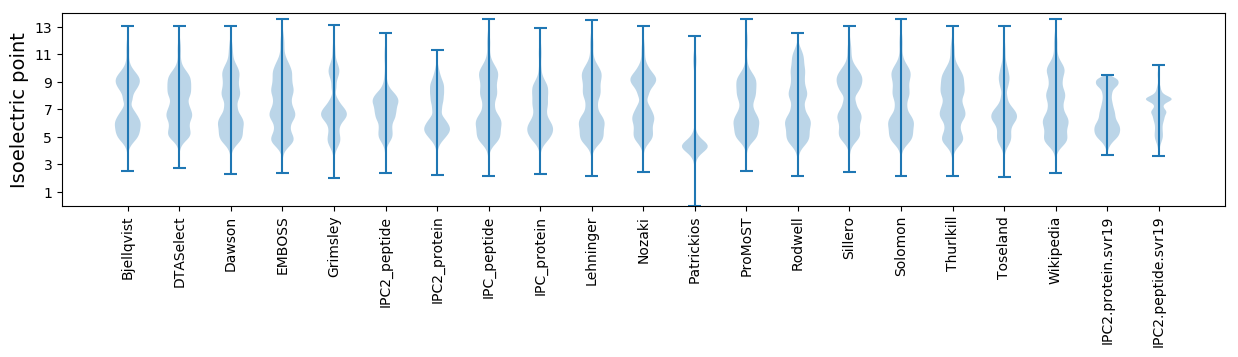
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2A2KBM4|A0A2A2KBM4_9BILA Uncharacterized protein OS=Diploscapter pachys OX=2018661 GN=WR25_03065 PE=4 SV=1
MM1 pKa = 7.69AAPAAAVVSWPRR13 pKa = 11.84PRR15 pKa = 11.84RR16 pKa = 11.84PRR18 pKa = 11.84RR19 pKa = 11.84ARR21 pKa = 11.84GSIALASAMRR31 pKa = 11.84AIARR35 pKa = 11.84APLAARR41 pKa = 11.84SASRR45 pKa = 11.84VPASAAASPATAATPGVASTSPPASTVPAPGSNSPMAPSRR85 pKa = 11.84TPRR88 pKa = 11.84TAALPRR94 pKa = 11.84PPASTAPAAA103 pKa = 3.99
MM1 pKa = 7.69AAPAAAVVSWPRR13 pKa = 11.84PRR15 pKa = 11.84RR16 pKa = 11.84PRR18 pKa = 11.84RR19 pKa = 11.84ARR21 pKa = 11.84GSIALASAMRR31 pKa = 11.84AIARR35 pKa = 11.84APLAARR41 pKa = 11.84SASRR45 pKa = 11.84VPASAAASPATAATPGVASTSPPASTVPAPGSNSPMAPSRR85 pKa = 11.84TPRR88 pKa = 11.84TAALPRR94 pKa = 11.84PPASTAPAAA103 pKa = 3.99
Molecular weight: 10.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
15129975 |
49 |
10228 |
445.2 |
49.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.323 ± 0.017 | 1.858 ± 0.011 |
5.576 ± 0.012 | 6.73 ± 0.019 |
3.993 ± 0.011 | 5.852 ± 0.013 |
2.375 ± 0.007 | 5.664 ± 0.011 |
5.91 ± 0.021 | 8.656 ± 0.019 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.539 ± 0.007 | 4.554 ± 0.011 |
5.19 ± 0.016 | 4.494 ± 0.013 |
5.968 ± 0.014 | 7.839 ± 0.019 |
5.484 ± 0.013 | 5.977 ± 0.012 |
1.086 ± 0.005 | 2.925 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
