
Hubei narna-like virus 18
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
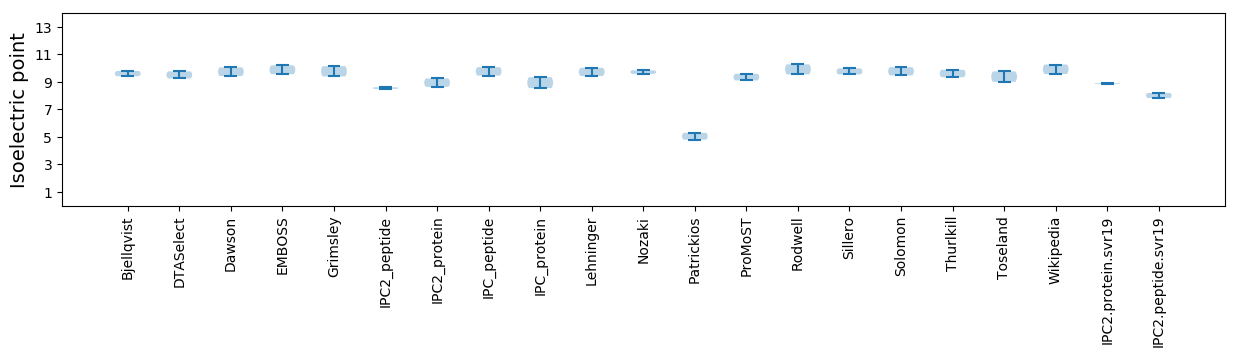
Average proteome isoelectric point is 8.87
Get precalculated fractions of proteins
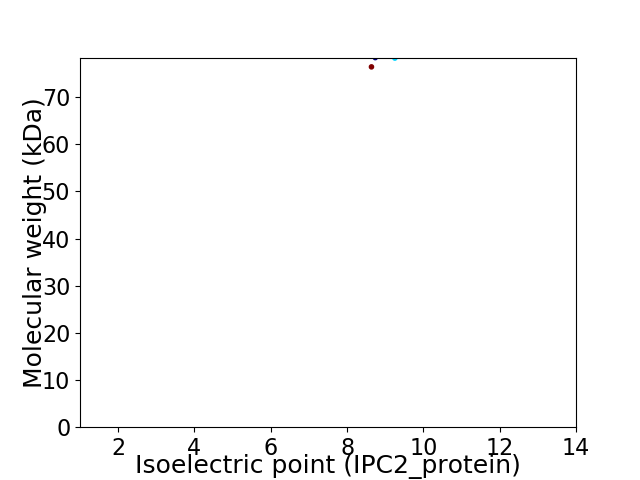
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KIC4|A0A1L3KIC4_9VIRU RNA-directed RNA polymerase OS=Hubei narna-like virus 18 OX=1922948 PE=4 SV=1
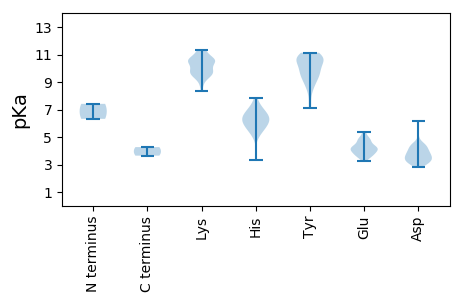
MM1 pKa = 6.32QWNWLRR7 pKa = 11.84MICGIISGIVEE18 pKa = 4.39KK19 pKa = 9.65HH20 pKa = 4.97TLPTITGKK28 pKa = 9.64IFRR31 pKa = 11.84GIRR34 pKa = 11.84VKK36 pKa = 10.81HH37 pKa = 5.46VDD39 pKa = 4.5RR40 pKa = 11.84FSLWKK45 pKa = 8.74ICNMLPIVLKK55 pKa = 10.72KK56 pKa = 9.5IARR59 pKa = 11.84ISNVEE64 pKa = 4.01LPHH67 pKa = 6.12SQLEE71 pKa = 4.27VMRR74 pKa = 11.84LVRR77 pKa = 11.84GCITNTRR84 pKa = 11.84SMLEE88 pKa = 3.51IRR90 pKa = 11.84VQLHH94 pKa = 4.98RR95 pKa = 11.84TPFIRR100 pKa = 11.84DD101 pKa = 3.2SAPAQFHH108 pKa = 6.1RR109 pKa = 11.84ACYY112 pKa = 9.07WDD114 pKa = 4.54AEE116 pKa = 4.28ATGYY120 pKa = 9.45PKK122 pKa = 10.62YY123 pKa = 10.74VSNRR127 pKa = 11.84TTDD130 pKa = 2.86RR131 pKa = 11.84TVRR134 pKa = 11.84HH135 pKa = 5.93LALPRR140 pKa = 11.84WVIRR144 pKa = 11.84RR145 pKa = 11.84PLGVPSAHH153 pKa = 6.45LPEE156 pKa = 4.84SATLIVQMDD165 pKa = 3.89EE166 pKa = 4.18TLKK169 pKa = 11.21GDD171 pKa = 4.43IIQIGYY177 pKa = 9.28RR178 pKa = 11.84RR179 pKa = 11.84NDD181 pKa = 4.02LLICDD186 pKa = 4.08YY187 pKa = 11.09KK188 pKa = 11.15VIEE191 pKa = 4.68SGSCQGVSFLGLPGNLHH208 pKa = 5.39GLFRR212 pKa = 11.84EE213 pKa = 4.77GTTVFGDD220 pKa = 3.2NRR222 pKa = 11.84FITVYY227 pKa = 10.76LKK229 pKa = 10.23TNLFHH234 pKa = 7.81DD235 pKa = 4.52SKK237 pKa = 10.58VLPWRR242 pKa = 11.84AEE244 pKa = 3.8HH245 pKa = 5.77TEE247 pKa = 4.08EE248 pKa = 5.19VISSDD253 pKa = 4.22DD254 pKa = 3.13IRR256 pKa = 11.84ATFVSRR262 pKa = 11.84LMDD265 pKa = 3.37TEE267 pKa = 4.89LVQKK271 pKa = 10.36RR272 pKa = 11.84EE273 pKa = 4.13CEE275 pKa = 3.83EE276 pKa = 4.09TQRR279 pKa = 11.84GSHH282 pKa = 6.55LNTATRR288 pKa = 11.84GHH290 pKa = 6.32GNSVRR295 pKa = 11.84IRR297 pKa = 11.84HH298 pKa = 5.88PNGCKK303 pKa = 9.98HH304 pKa = 5.85PFKK307 pKa = 11.14VVFNPFRR314 pKa = 11.84EE315 pKa = 4.41VKK317 pKa = 10.22RR318 pKa = 11.84IEE320 pKa = 4.21NSSPEE325 pKa = 4.29CLCVLVGYY333 pKa = 7.08VVRR336 pKa = 11.84RR337 pKa = 11.84VGQIRR342 pKa = 11.84RR343 pKa = 11.84YY344 pKa = 9.1DD345 pKa = 3.62CTVSVQRR352 pKa = 11.84TADD355 pKa = 3.93HH356 pKa = 6.28ILEE359 pKa = 4.3LLRR362 pKa = 11.84AYY364 pKa = 9.75ILIEE368 pKa = 3.27VHH370 pKa = 5.29IRR372 pKa = 11.84VRR374 pKa = 11.84LEE376 pKa = 3.53YY377 pKa = 10.64GPQLSADD384 pKa = 4.2DD385 pKa = 3.69ITRR388 pKa = 11.84KK389 pKa = 9.42EE390 pKa = 3.97SCGWEE395 pKa = 3.75LAEE398 pKa = 4.13YY399 pKa = 9.43TGLKK403 pKa = 9.72PGFRR407 pKa = 11.84NGEE410 pKa = 4.12NVHH413 pKa = 6.39VISGDD418 pKa = 3.6LFVSIDD424 pKa = 3.39SSFDD428 pKa = 3.43DD429 pKa = 4.87SIVKK433 pKa = 9.72VLLMTIKK440 pKa = 10.64VLAQYY445 pKa = 11.01DD446 pKa = 3.51GTLRR450 pKa = 11.84HH451 pKa = 5.79NSVGIVHH458 pKa = 6.92KK459 pKa = 10.6VSQSTTRR466 pKa = 11.84ASRR469 pKa = 11.84NISASRR475 pKa = 11.84SFRR478 pKa = 11.84WMEE481 pKa = 3.67GHH483 pKa = 7.6PLTQAVFTLVNIITSVAINLVCQTNIQLWSAGHH516 pKa = 6.02NAPVFPEE523 pKa = 3.48RR524 pKa = 11.84LVYY527 pKa = 10.93NKK529 pKa = 9.31FVPNRR534 pKa = 11.84EE535 pKa = 4.06HH536 pKa = 6.67SCYY539 pKa = 10.53ACKK542 pKa = 10.55LCTRR546 pKa = 11.84LCMTAEE552 pKa = 3.85AAKK555 pKa = 9.92PVSKK559 pKa = 10.49CSWYY563 pKa = 8.69IPRR566 pKa = 11.84VIRR569 pKa = 11.84LLSSYY574 pKa = 10.8RR575 pKa = 11.84PDD577 pKa = 3.16TVVEE581 pKa = 4.2GQRR584 pKa = 11.84FNLLADD590 pKa = 3.55ILLTTGTTTGSNKK603 pKa = 9.59PPRR606 pKa = 11.84KK607 pKa = 9.38HH608 pKa = 6.72LSGDD612 pKa = 3.83QIRR615 pKa = 11.84EE616 pKa = 3.74WSPVYY621 pKa = 10.63GFYY624 pKa = 10.7NDD626 pKa = 3.41SHH628 pKa = 6.23TLPDD632 pKa = 3.64VGDD635 pKa = 3.85KK636 pKa = 11.14LSLSFIRR643 pKa = 11.84QGRR646 pKa = 11.84HH647 pKa = 3.31WQLVPVTCPDD657 pKa = 3.0NTGDD661 pKa = 4.14DD662 pKa = 3.92RR663 pKa = 11.84PRR665 pKa = 11.84QDD667 pKa = 2.96ILRR670 pKa = 4.27
MM1 pKa = 6.32QWNWLRR7 pKa = 11.84MICGIISGIVEE18 pKa = 4.39KK19 pKa = 9.65HH20 pKa = 4.97TLPTITGKK28 pKa = 9.64IFRR31 pKa = 11.84GIRR34 pKa = 11.84VKK36 pKa = 10.81HH37 pKa = 5.46VDD39 pKa = 4.5RR40 pKa = 11.84FSLWKK45 pKa = 8.74ICNMLPIVLKK55 pKa = 10.72KK56 pKa = 9.5IARR59 pKa = 11.84ISNVEE64 pKa = 4.01LPHH67 pKa = 6.12SQLEE71 pKa = 4.27VMRR74 pKa = 11.84LVRR77 pKa = 11.84GCITNTRR84 pKa = 11.84SMLEE88 pKa = 3.51IRR90 pKa = 11.84VQLHH94 pKa = 4.98RR95 pKa = 11.84TPFIRR100 pKa = 11.84DD101 pKa = 3.2SAPAQFHH108 pKa = 6.1RR109 pKa = 11.84ACYY112 pKa = 9.07WDD114 pKa = 4.54AEE116 pKa = 4.28ATGYY120 pKa = 9.45PKK122 pKa = 10.62YY123 pKa = 10.74VSNRR127 pKa = 11.84TTDD130 pKa = 2.86RR131 pKa = 11.84TVRR134 pKa = 11.84HH135 pKa = 5.93LALPRR140 pKa = 11.84WVIRR144 pKa = 11.84RR145 pKa = 11.84PLGVPSAHH153 pKa = 6.45LPEE156 pKa = 4.84SATLIVQMDD165 pKa = 3.89EE166 pKa = 4.18TLKK169 pKa = 11.21GDD171 pKa = 4.43IIQIGYY177 pKa = 9.28RR178 pKa = 11.84RR179 pKa = 11.84NDD181 pKa = 4.02LLICDD186 pKa = 4.08YY187 pKa = 11.09KK188 pKa = 11.15VIEE191 pKa = 4.68SGSCQGVSFLGLPGNLHH208 pKa = 5.39GLFRR212 pKa = 11.84EE213 pKa = 4.77GTTVFGDD220 pKa = 3.2NRR222 pKa = 11.84FITVYY227 pKa = 10.76LKK229 pKa = 10.23TNLFHH234 pKa = 7.81DD235 pKa = 4.52SKK237 pKa = 10.58VLPWRR242 pKa = 11.84AEE244 pKa = 3.8HH245 pKa = 5.77TEE247 pKa = 4.08EE248 pKa = 5.19VISSDD253 pKa = 4.22DD254 pKa = 3.13IRR256 pKa = 11.84ATFVSRR262 pKa = 11.84LMDD265 pKa = 3.37TEE267 pKa = 4.89LVQKK271 pKa = 10.36RR272 pKa = 11.84EE273 pKa = 4.13CEE275 pKa = 3.83EE276 pKa = 4.09TQRR279 pKa = 11.84GSHH282 pKa = 6.55LNTATRR288 pKa = 11.84GHH290 pKa = 6.32GNSVRR295 pKa = 11.84IRR297 pKa = 11.84HH298 pKa = 5.88PNGCKK303 pKa = 9.98HH304 pKa = 5.85PFKK307 pKa = 11.14VVFNPFRR314 pKa = 11.84EE315 pKa = 4.41VKK317 pKa = 10.22RR318 pKa = 11.84IEE320 pKa = 4.21NSSPEE325 pKa = 4.29CLCVLVGYY333 pKa = 7.08VVRR336 pKa = 11.84RR337 pKa = 11.84VGQIRR342 pKa = 11.84RR343 pKa = 11.84YY344 pKa = 9.1DD345 pKa = 3.62CTVSVQRR352 pKa = 11.84TADD355 pKa = 3.93HH356 pKa = 6.28ILEE359 pKa = 4.3LLRR362 pKa = 11.84AYY364 pKa = 9.75ILIEE368 pKa = 3.27VHH370 pKa = 5.29IRR372 pKa = 11.84VRR374 pKa = 11.84LEE376 pKa = 3.53YY377 pKa = 10.64GPQLSADD384 pKa = 4.2DD385 pKa = 3.69ITRR388 pKa = 11.84KK389 pKa = 9.42EE390 pKa = 3.97SCGWEE395 pKa = 3.75LAEE398 pKa = 4.13YY399 pKa = 9.43TGLKK403 pKa = 9.72PGFRR407 pKa = 11.84NGEE410 pKa = 4.12NVHH413 pKa = 6.39VISGDD418 pKa = 3.6LFVSIDD424 pKa = 3.39SSFDD428 pKa = 3.43DD429 pKa = 4.87SIVKK433 pKa = 9.72VLLMTIKK440 pKa = 10.64VLAQYY445 pKa = 11.01DD446 pKa = 3.51GTLRR450 pKa = 11.84HH451 pKa = 5.79NSVGIVHH458 pKa = 6.92KK459 pKa = 10.6VSQSTTRR466 pKa = 11.84ASRR469 pKa = 11.84NISASRR475 pKa = 11.84SFRR478 pKa = 11.84WMEE481 pKa = 3.67GHH483 pKa = 7.6PLTQAVFTLVNIITSVAINLVCQTNIQLWSAGHH516 pKa = 6.02NAPVFPEE523 pKa = 3.48RR524 pKa = 11.84LVYY527 pKa = 10.93NKK529 pKa = 9.31FVPNRR534 pKa = 11.84EE535 pKa = 4.06HH536 pKa = 6.67SCYY539 pKa = 10.53ACKK542 pKa = 10.55LCTRR546 pKa = 11.84LCMTAEE552 pKa = 3.85AAKK555 pKa = 9.92PVSKK559 pKa = 10.49CSWYY563 pKa = 8.69IPRR566 pKa = 11.84VIRR569 pKa = 11.84LLSSYY574 pKa = 10.8RR575 pKa = 11.84PDD577 pKa = 3.16TVVEE581 pKa = 4.2GQRR584 pKa = 11.84FNLLADD590 pKa = 3.55ILLTTGTTTGSNKK603 pKa = 9.59PPRR606 pKa = 11.84KK607 pKa = 9.38HH608 pKa = 6.72LSGDD612 pKa = 3.83QIRR615 pKa = 11.84EE616 pKa = 3.74WSPVYY621 pKa = 10.63GFYY624 pKa = 10.7NDD626 pKa = 3.41SHH628 pKa = 6.23TLPDD632 pKa = 3.64VGDD635 pKa = 3.85KK636 pKa = 11.14LSLSFIRR643 pKa = 11.84QGRR646 pKa = 11.84HH647 pKa = 3.31WQLVPVTCPDD657 pKa = 3.0NTGDD661 pKa = 4.14DD662 pKa = 3.92RR663 pKa = 11.84PRR665 pKa = 11.84QDD667 pKa = 2.96ILRR670 pKa = 4.27
Molecular weight: 76.4 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KIC4|A0A1L3KIC4_9VIRU RNA-directed RNA polymerase OS=Hubei narna-like virus 18 OX=1922948 PE=4 SV=1
MM1 pKa = 7.41AVVIEE6 pKa = 4.83AIHH9 pKa = 6.24RR10 pKa = 11.84RR11 pKa = 11.84PFTNLVPAKK20 pKa = 9.63MFAGWLIRR28 pKa = 11.84ASRR31 pKa = 11.84GPGGQKK37 pKa = 9.71YY38 pKa = 8.93VCQQIKK44 pKa = 10.52SLAFDD49 pKa = 3.5YY50 pKa = 10.63RR51 pKa = 11.84VWSITAKK58 pKa = 10.1KK59 pKa = 9.31PNYY62 pKa = 10.22SGDD65 pKa = 3.13IPRR68 pKa = 11.84AFRR71 pKa = 11.84NWLSSLCCHH80 pKa = 6.91AEE82 pKa = 3.74TRR84 pKa = 11.84AQFARR89 pKa = 11.84VARR92 pKa = 11.84MLPVGNKK99 pKa = 9.74LVIDD103 pKa = 3.62QALRR107 pKa = 11.84KK108 pKa = 8.88HH109 pKa = 6.2RR110 pKa = 11.84SVVTRR115 pKa = 11.84RR116 pKa = 11.84PKK118 pKa = 10.67LDD120 pKa = 3.1IGLAHH125 pKa = 6.76QIYY128 pKa = 10.07GYY130 pKa = 10.94ARR132 pKa = 11.84DD133 pKa = 3.93YY134 pKa = 10.31IHH136 pKa = 6.78EE137 pKa = 4.61RR138 pKa = 11.84KK139 pKa = 9.66HH140 pKa = 5.74SLSEE144 pKa = 3.82RR145 pKa = 11.84VSFHH149 pKa = 5.65PTEE152 pKa = 4.49GTASRR157 pKa = 11.84DD158 pKa = 3.55VPASAGGRR166 pKa = 11.84LRR168 pKa = 11.84DD169 pKa = 3.97LVDD172 pKa = 3.27NAHH175 pKa = 6.6RR176 pKa = 11.84VMSQRR181 pKa = 11.84PIVLSEE187 pKa = 3.49YY188 pKa = 10.44LYY190 pKa = 10.35SHH192 pKa = 7.15KK193 pKa = 10.7KK194 pKa = 10.01YY195 pKa = 10.58FDD197 pKa = 3.8DD198 pKa = 6.17AIVEE202 pKa = 4.08AAIYY206 pKa = 9.7TDD208 pKa = 3.96EE209 pKa = 4.79KK210 pKa = 10.69VSTYY214 pKa = 10.92NVDD217 pKa = 3.08VLAIPEE223 pKa = 4.49PGFKK227 pKa = 10.65ARR229 pKa = 11.84VLCKK233 pKa = 10.42FPATALLAGDD243 pKa = 4.33IIRR246 pKa = 11.84RR247 pKa = 11.84QLWPIFEE254 pKa = 5.21SDD256 pKa = 3.22PNMDD260 pKa = 3.65FDD262 pKa = 4.43QDD264 pKa = 3.13IRR266 pKa = 11.84SEE268 pKa = 3.99KK269 pKa = 8.33FQNVIRR275 pKa = 11.84RR276 pKa = 11.84SLNRR280 pKa = 11.84DD281 pKa = 3.06GTIVSSDD288 pKa = 3.23LSNATDD294 pKa = 4.32YY295 pKa = 11.13IPHH298 pKa = 7.19EE299 pKa = 4.17YY300 pKa = 10.76AKK302 pKa = 10.74ALWAGILDD310 pKa = 3.99AFDD313 pKa = 3.81FPEE316 pKa = 3.9WVEE319 pKa = 4.11NYY321 pKa = 9.73LEE323 pKa = 4.93RR324 pKa = 11.84MFAPIRR330 pKa = 11.84MSYY333 pKa = 9.88PDD335 pKa = 3.82GVTVTSCRR343 pKa = 11.84GIQMGTPLSFLTLSLLHH360 pKa = 6.38KK361 pKa = 10.41FCVHH365 pKa = 6.4KK366 pKa = 10.61SGHH369 pKa = 5.4EE370 pKa = 3.66RR371 pKa = 11.84SPYY374 pKa = 9.85IIRR377 pKa = 11.84GDD379 pKa = 3.66DD380 pKa = 3.47LLGVFSSPRR389 pKa = 11.84QYY391 pKa = 9.97LTVMEE396 pKa = 4.84EE397 pKa = 3.47IGFKK401 pKa = 10.28INRR404 pKa = 11.84DD405 pKa = 3.03KK406 pKa = 11.31TVISKK411 pKa = 10.82DD412 pKa = 2.81GGTFAEE418 pKa = 4.4QTVKK422 pKa = 9.39VTWKK426 pKa = 10.53AKK428 pKa = 10.13EE429 pKa = 3.92RR430 pKa = 11.84DD431 pKa = 3.45PLARR435 pKa = 11.84PTLYY439 pKa = 10.96DD440 pKa = 5.51FIVTDD445 pKa = 3.57QKK447 pKa = 11.34VVSSITNLDD456 pKa = 4.41DD457 pKa = 4.2IPFKK461 pKa = 11.22GLIHH465 pKa = 7.18LDD467 pKa = 3.45NKK469 pKa = 10.39GGRR472 pKa = 11.84LRR474 pKa = 11.84QVGRR478 pKa = 11.84WYY480 pKa = 10.51AQWSPYY486 pKa = 9.43YY487 pKa = 9.36PPRR490 pKa = 11.84KK491 pKa = 9.83GKK493 pKa = 9.16VAYY496 pKa = 9.76RR497 pKa = 11.84AIRR500 pKa = 11.84RR501 pKa = 11.84TIGNVLRR508 pKa = 11.84IARR511 pKa = 11.84SLRR514 pKa = 11.84IPITCPMEE522 pKa = 4.79LGGCGIPNKK531 pKa = 10.05RR532 pKa = 11.84GTMQLDD538 pKa = 3.3ANFQHH543 pKa = 7.26RR544 pKa = 11.84SRR546 pKa = 11.84VGYY549 pKa = 9.28AASHH553 pKa = 6.28EE554 pKa = 4.42SHH556 pKa = 6.59NFQLAVRR563 pKa = 11.84KK564 pKa = 9.8LDD566 pKa = 3.46IGNASDD572 pKa = 4.23LLEE575 pKa = 5.36DD576 pKa = 4.03YY577 pKa = 9.75RR578 pKa = 11.84QHH580 pKa = 6.83VADD583 pKa = 4.37LPKK586 pKa = 10.72GKK588 pKa = 9.53SVYY591 pKa = 10.15MFDD594 pKa = 4.45PYY596 pKa = 11.07SSEE599 pKa = 4.32DD600 pKa = 3.36FSGYY604 pKa = 8.49RR605 pKa = 11.84RR606 pKa = 11.84KK607 pKa = 10.47RR608 pKa = 11.84MLLNNTGNYY617 pKa = 8.69SAYY620 pKa = 8.24HH621 pKa = 5.55TKK623 pKa = 10.04PVPLHH628 pKa = 5.84RR629 pKa = 11.84WLAVFSRR636 pKa = 11.84CKK638 pKa = 9.8EE639 pKa = 3.7NRR641 pKa = 11.84ARR643 pKa = 11.84WVPSRR648 pKa = 11.84RR649 pKa = 11.84CSGSDD654 pKa = 3.22LRR656 pKa = 11.84RR657 pKa = 11.84LIDD660 pKa = 4.41RR661 pKa = 11.84IKK663 pKa = 9.24MLSPIYY669 pKa = 10.6VPYY672 pKa = 7.48TTPNAQILSIRR683 pKa = 11.84II684 pKa = 3.65
MM1 pKa = 7.41AVVIEE6 pKa = 4.83AIHH9 pKa = 6.24RR10 pKa = 11.84RR11 pKa = 11.84PFTNLVPAKK20 pKa = 9.63MFAGWLIRR28 pKa = 11.84ASRR31 pKa = 11.84GPGGQKK37 pKa = 9.71YY38 pKa = 8.93VCQQIKK44 pKa = 10.52SLAFDD49 pKa = 3.5YY50 pKa = 10.63RR51 pKa = 11.84VWSITAKK58 pKa = 10.1KK59 pKa = 9.31PNYY62 pKa = 10.22SGDD65 pKa = 3.13IPRR68 pKa = 11.84AFRR71 pKa = 11.84NWLSSLCCHH80 pKa = 6.91AEE82 pKa = 3.74TRR84 pKa = 11.84AQFARR89 pKa = 11.84VARR92 pKa = 11.84MLPVGNKK99 pKa = 9.74LVIDD103 pKa = 3.62QALRR107 pKa = 11.84KK108 pKa = 8.88HH109 pKa = 6.2RR110 pKa = 11.84SVVTRR115 pKa = 11.84RR116 pKa = 11.84PKK118 pKa = 10.67LDD120 pKa = 3.1IGLAHH125 pKa = 6.76QIYY128 pKa = 10.07GYY130 pKa = 10.94ARR132 pKa = 11.84DD133 pKa = 3.93YY134 pKa = 10.31IHH136 pKa = 6.78EE137 pKa = 4.61RR138 pKa = 11.84KK139 pKa = 9.66HH140 pKa = 5.74SLSEE144 pKa = 3.82RR145 pKa = 11.84VSFHH149 pKa = 5.65PTEE152 pKa = 4.49GTASRR157 pKa = 11.84DD158 pKa = 3.55VPASAGGRR166 pKa = 11.84LRR168 pKa = 11.84DD169 pKa = 3.97LVDD172 pKa = 3.27NAHH175 pKa = 6.6RR176 pKa = 11.84VMSQRR181 pKa = 11.84PIVLSEE187 pKa = 3.49YY188 pKa = 10.44LYY190 pKa = 10.35SHH192 pKa = 7.15KK193 pKa = 10.7KK194 pKa = 10.01YY195 pKa = 10.58FDD197 pKa = 3.8DD198 pKa = 6.17AIVEE202 pKa = 4.08AAIYY206 pKa = 9.7TDD208 pKa = 3.96EE209 pKa = 4.79KK210 pKa = 10.69VSTYY214 pKa = 10.92NVDD217 pKa = 3.08VLAIPEE223 pKa = 4.49PGFKK227 pKa = 10.65ARR229 pKa = 11.84VLCKK233 pKa = 10.42FPATALLAGDD243 pKa = 4.33IIRR246 pKa = 11.84RR247 pKa = 11.84QLWPIFEE254 pKa = 5.21SDD256 pKa = 3.22PNMDD260 pKa = 3.65FDD262 pKa = 4.43QDD264 pKa = 3.13IRR266 pKa = 11.84SEE268 pKa = 3.99KK269 pKa = 8.33FQNVIRR275 pKa = 11.84RR276 pKa = 11.84SLNRR280 pKa = 11.84DD281 pKa = 3.06GTIVSSDD288 pKa = 3.23LSNATDD294 pKa = 4.32YY295 pKa = 11.13IPHH298 pKa = 7.19EE299 pKa = 4.17YY300 pKa = 10.76AKK302 pKa = 10.74ALWAGILDD310 pKa = 3.99AFDD313 pKa = 3.81FPEE316 pKa = 3.9WVEE319 pKa = 4.11NYY321 pKa = 9.73LEE323 pKa = 4.93RR324 pKa = 11.84MFAPIRR330 pKa = 11.84MSYY333 pKa = 9.88PDD335 pKa = 3.82GVTVTSCRR343 pKa = 11.84GIQMGTPLSFLTLSLLHH360 pKa = 6.38KK361 pKa = 10.41FCVHH365 pKa = 6.4KK366 pKa = 10.61SGHH369 pKa = 5.4EE370 pKa = 3.66RR371 pKa = 11.84SPYY374 pKa = 9.85IIRR377 pKa = 11.84GDD379 pKa = 3.66DD380 pKa = 3.47LLGVFSSPRR389 pKa = 11.84QYY391 pKa = 9.97LTVMEE396 pKa = 4.84EE397 pKa = 3.47IGFKK401 pKa = 10.28INRR404 pKa = 11.84DD405 pKa = 3.03KK406 pKa = 11.31TVISKK411 pKa = 10.82DD412 pKa = 2.81GGTFAEE418 pKa = 4.4QTVKK422 pKa = 9.39VTWKK426 pKa = 10.53AKK428 pKa = 10.13EE429 pKa = 3.92RR430 pKa = 11.84DD431 pKa = 3.45PLARR435 pKa = 11.84PTLYY439 pKa = 10.96DD440 pKa = 5.51FIVTDD445 pKa = 3.57QKK447 pKa = 11.34VVSSITNLDD456 pKa = 4.41DD457 pKa = 4.2IPFKK461 pKa = 11.22GLIHH465 pKa = 7.18LDD467 pKa = 3.45NKK469 pKa = 10.39GGRR472 pKa = 11.84LRR474 pKa = 11.84QVGRR478 pKa = 11.84WYY480 pKa = 10.51AQWSPYY486 pKa = 9.43YY487 pKa = 9.36PPRR490 pKa = 11.84KK491 pKa = 9.83GKK493 pKa = 9.16VAYY496 pKa = 9.76RR497 pKa = 11.84AIRR500 pKa = 11.84RR501 pKa = 11.84TIGNVLRR508 pKa = 11.84IARR511 pKa = 11.84SLRR514 pKa = 11.84IPITCPMEE522 pKa = 4.79LGGCGIPNKK531 pKa = 10.05RR532 pKa = 11.84GTMQLDD538 pKa = 3.3ANFQHH543 pKa = 7.26RR544 pKa = 11.84SRR546 pKa = 11.84VGYY549 pKa = 9.28AASHH553 pKa = 6.28EE554 pKa = 4.42SHH556 pKa = 6.59NFQLAVRR563 pKa = 11.84KK564 pKa = 9.8LDD566 pKa = 3.46IGNASDD572 pKa = 4.23LLEE575 pKa = 5.36DD576 pKa = 4.03YY577 pKa = 9.75RR578 pKa = 11.84QHH580 pKa = 6.83VADD583 pKa = 4.37LPKK586 pKa = 10.72GKK588 pKa = 9.53SVYY591 pKa = 10.15MFDD594 pKa = 4.45PYY596 pKa = 11.07SSEE599 pKa = 4.32DD600 pKa = 3.36FSGYY604 pKa = 8.49RR605 pKa = 11.84RR606 pKa = 11.84KK607 pKa = 10.47RR608 pKa = 11.84MLLNNTGNYY617 pKa = 8.69SAYY620 pKa = 8.24HH621 pKa = 5.55TKK623 pKa = 10.04PVPLHH628 pKa = 5.84RR629 pKa = 11.84WLAVFSRR636 pKa = 11.84CKK638 pKa = 9.8EE639 pKa = 3.7NRR641 pKa = 11.84ARR643 pKa = 11.84WVPSRR648 pKa = 11.84RR649 pKa = 11.84CSGSDD654 pKa = 3.22LRR656 pKa = 11.84RR657 pKa = 11.84LIDD660 pKa = 4.41RR661 pKa = 11.84IKK663 pKa = 9.24MLSPIYY669 pKa = 10.6VPYY672 pKa = 7.48TTPNAQILSIRR683 pKa = 11.84II684 pKa = 3.65
Molecular weight: 78.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1354 |
670 |
684 |
677.0 |
77.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.761 ± 1.13 | 2.142 ± 0.496 |
5.465 ± 0.492 | 4.21 ± 0.405 |
3.693 ± 0.186 | 5.908 ± 0.044 |
3.323 ± 0.291 | 6.942 ± 0.158 |
4.727 ± 0.498 | 8.715 ± 0.385 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.773 ± 0.2 | 3.84 ± 0.242 |
5.244 ± 0.334 | 3.176 ± 0.077 |
9.38 ± 0.197 | 7.238 ± 0.053 |
5.539 ± 0.841 | 7.607 ± 0.75 |
1.699 ± 0.066 | 3.619 ± 0.666 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
