
Agrilactobacillus composti DSM 18527 = JCM 14202
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Agrilactobacillus; Agrilactobacillus composti
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
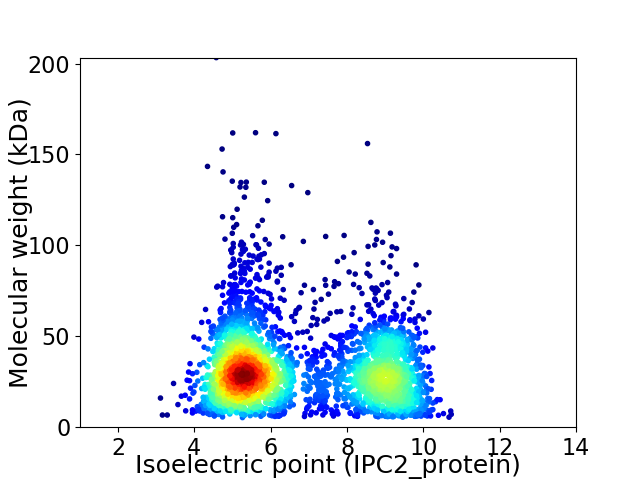
Virtual 2D-PAGE plot for 3260 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0R1Y2M9|A0A0R1Y2M9_9LACO Glycosyl transferase 2 family protein OS=Agrilactobacillus composti DSM 18527 = JCM 14202 OX=1423734 GN=FC83_GL003004 PE=4 SV=1
MM1 pKa = 7.6ASVKK5 pKa = 10.23IIEE8 pKa = 4.35TSVASGNTGGDD19 pKa = 3.38MYY21 pKa = 11.1DD22 pKa = 4.02SPNGTYY28 pKa = 10.06IKK30 pKa = 10.73ALALNTDD37 pKa = 2.96WEE39 pKa = 4.54YY40 pKa = 11.73FDD42 pKa = 3.89TQTVDD47 pKa = 3.12GQIWYY52 pKa = 9.58DD53 pKa = 3.49LGGDD57 pKa = 3.38QWISEE62 pKa = 4.37IYY64 pKa = 10.0AAPNINDD71 pKa = 3.69LEE73 pKa = 4.45VRR75 pKa = 11.84SGTAKK80 pKa = 10.59VADD83 pKa = 3.98FTTATVYY90 pKa = 10.19YY91 pKa = 9.85SPNGSEE97 pKa = 4.0NTQDD101 pKa = 3.67NPTPLPEE108 pKa = 4.22NSEE111 pKa = 3.8WAYY114 pKa = 11.48NKK116 pKa = 10.4AATDD120 pKa = 3.67SEE122 pKa = 4.39GTIWFDD128 pKa = 3.06VGRR131 pKa = 11.84SQWVTTQEE139 pKa = 4.44VVDD142 pKa = 4.36EE143 pKa = 4.57DD144 pKa = 3.83AGGDD148 pKa = 3.91IEE150 pKa = 4.48WSGSGIFQYY159 pKa = 8.49PTYY162 pKa = 10.89SDD164 pKa = 3.42GGINADD170 pKa = 4.68MIRR173 pKa = 11.84QAASDD178 pKa = 4.06MYY180 pKa = 10.84TSVSDD185 pKa = 3.91DD186 pKa = 3.67QINTIMAVAEE196 pKa = 4.3HH197 pKa = 6.69EE198 pKa = 4.57SGNNSNKK205 pKa = 9.26INNWDD210 pKa = 3.41KK211 pKa = 10.94NAQEE215 pKa = 4.12GHH217 pKa = 6.38PSKK220 pKa = 11.03GVLQFVGYY228 pKa = 8.27TFDD231 pKa = 3.86HH232 pKa = 5.7YY233 pKa = 11.17HH234 pKa = 5.87VANHH238 pKa = 6.33DD239 pKa = 4.62NIWSSMDD246 pKa = 3.15QLYY249 pKa = 11.26ALFNDD254 pKa = 4.11ATWASDD260 pKa = 2.83ISLGGWAPNGPVRR273 pKa = 11.84FPAA276 pKa = 4.36
MM1 pKa = 7.6ASVKK5 pKa = 10.23IIEE8 pKa = 4.35TSVASGNTGGDD19 pKa = 3.38MYY21 pKa = 11.1DD22 pKa = 4.02SPNGTYY28 pKa = 10.06IKK30 pKa = 10.73ALALNTDD37 pKa = 2.96WEE39 pKa = 4.54YY40 pKa = 11.73FDD42 pKa = 3.89TQTVDD47 pKa = 3.12GQIWYY52 pKa = 9.58DD53 pKa = 3.49LGGDD57 pKa = 3.38QWISEE62 pKa = 4.37IYY64 pKa = 10.0AAPNINDD71 pKa = 3.69LEE73 pKa = 4.45VRR75 pKa = 11.84SGTAKK80 pKa = 10.59VADD83 pKa = 3.98FTTATVYY90 pKa = 10.19YY91 pKa = 9.85SPNGSEE97 pKa = 4.0NTQDD101 pKa = 3.67NPTPLPEE108 pKa = 4.22NSEE111 pKa = 3.8WAYY114 pKa = 11.48NKK116 pKa = 10.4AATDD120 pKa = 3.67SEE122 pKa = 4.39GTIWFDD128 pKa = 3.06VGRR131 pKa = 11.84SQWVTTQEE139 pKa = 4.44VVDD142 pKa = 4.36EE143 pKa = 4.57DD144 pKa = 3.83AGGDD148 pKa = 3.91IEE150 pKa = 4.48WSGSGIFQYY159 pKa = 8.49PTYY162 pKa = 10.89SDD164 pKa = 3.42GGINADD170 pKa = 4.68MIRR173 pKa = 11.84QAASDD178 pKa = 4.06MYY180 pKa = 10.84TSVSDD185 pKa = 3.91DD186 pKa = 3.67QINTIMAVAEE196 pKa = 4.3HH197 pKa = 6.69EE198 pKa = 4.57SGNNSNKK205 pKa = 9.26INNWDD210 pKa = 3.41KK211 pKa = 10.94NAQEE215 pKa = 4.12GHH217 pKa = 6.38PSKK220 pKa = 11.03GVLQFVGYY228 pKa = 8.27TFDD231 pKa = 3.86HH232 pKa = 5.7YY233 pKa = 11.17HH234 pKa = 5.87VANHH238 pKa = 6.33DD239 pKa = 4.62NIWSSMDD246 pKa = 3.15QLYY249 pKa = 11.26ALFNDD254 pKa = 4.11ATWASDD260 pKa = 2.83ISLGGWAPNGPVRR273 pKa = 11.84FPAA276 pKa = 4.36
Molecular weight: 30.33 kDa
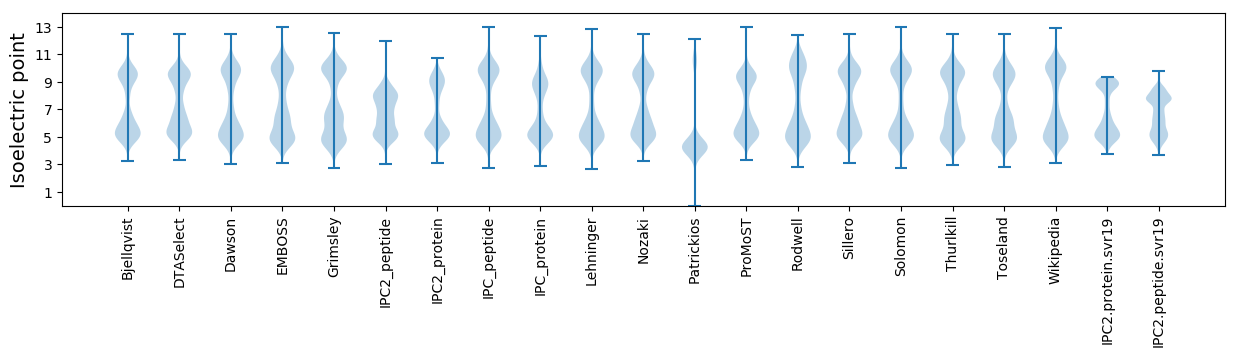
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|X0QHW8|X0QHW8_9LACO Uncharacterized protein OS=Agrilactobacillus composti DSM 18527 = JCM 14202 OX=1423734 GN=FC83_GL001830 PE=4 SV=1
MM1 pKa = 7.2FHH3 pKa = 7.3LIWVLIIGALIGVIAGAITSRR24 pKa = 11.84GMPFGWIGNILAGLIGAWLGQRR46 pKa = 11.84LLGTWGPSLAGMALIPAIIGAIILVLVVSLILGATTKK83 pKa = 10.41RR84 pKa = 11.84RR85 pKa = 11.84AA86 pKa = 3.17
MM1 pKa = 7.2FHH3 pKa = 7.3LIWVLIIGALIGVIAGAITSRR24 pKa = 11.84GMPFGWIGNILAGLIGAWLGQRR46 pKa = 11.84LLGTWGPSLAGMALIPAIIGAIILVLVVSLILGATTKK83 pKa = 10.41RR84 pKa = 11.84RR85 pKa = 11.84AA86 pKa = 3.17
Molecular weight: 8.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1004349 |
49 |
1815 |
308.1 |
34.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.184 ± 0.047 | 0.534 ± 0.012 |
5.667 ± 0.043 | 4.379 ± 0.036 |
4.356 ± 0.038 | 6.881 ± 0.035 |
2.124 ± 0.02 | 7.004 ± 0.041 |
5.849 ± 0.037 | 10.307 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.505 ± 0.021 | 4.591 ± 0.033 |
4.046 ± 0.029 | 5.268 ± 0.046 |
3.754 ± 0.031 | 5.379 ± 0.037 |
6.59 ± 0.048 | 6.969 ± 0.032 |
1.062 ± 0.017 | 3.539 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
