
Firmicutes bacterium CAG:94
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; environmental samples
Average proteome isoelectric point is 6.19
Get precalculated fractions of proteins
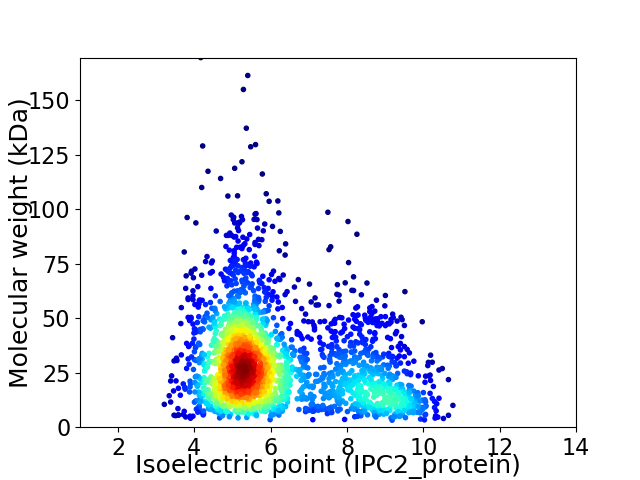
Virtual 2D-PAGE plot for 2410 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R6ZL02|R6ZL02_9FIRM 4-hydroxy-tetrahydrodipicolinate reductase OS=Firmicutes bacterium CAG:94 OX=1262989 GN=dapB PE=3 SV=1
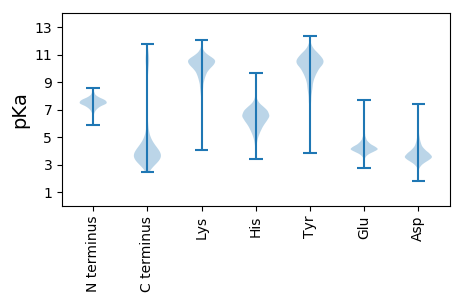
MM1 pKa = 7.02MRR3 pKa = 11.84IGSAIRR9 pKa = 11.84RR10 pKa = 11.84VLALALCVLLGCGAFAGCSQVDD32 pKa = 4.08DD33 pKa = 3.77IVSNVTAAGVFPVEE47 pKa = 4.4VNGVTISSRR56 pKa = 11.84PSRR59 pKa = 11.84VVVLSPSLADD69 pKa = 3.59VILALGCEE77 pKa = 4.17TQLAGASEE85 pKa = 4.96GCTQSALDD93 pKa = 3.84EE94 pKa = 4.44LEE96 pKa = 4.39KK97 pKa = 11.22VSADD101 pKa = 3.28SVEE104 pKa = 4.9AIQALQPDD112 pKa = 5.12LVLVDD117 pKa = 4.67PNSSGAQSALEE128 pKa = 3.99EE129 pKa = 4.39AGVTVLSVEE138 pKa = 4.24PATDD142 pKa = 3.21RR143 pKa = 11.84SDD145 pKa = 3.72FEE147 pKa = 4.45RR148 pKa = 11.84LYY150 pKa = 10.74SQVSSALLGGEE161 pKa = 3.87AGYY164 pKa = 11.03DD165 pKa = 3.35EE166 pKa = 6.1GIATAQDD173 pKa = 3.11IFITLDD179 pKa = 3.47NINRR183 pKa = 11.84IVPKK187 pKa = 9.9DD188 pKa = 3.78TITTACYY195 pKa = 10.13LYY197 pKa = 10.78DD198 pKa = 4.4LDD200 pKa = 4.67GSAVTGDD207 pKa = 3.71MFGSTIMSYY216 pKa = 11.07AGVTNVFEE224 pKa = 4.3SLEE227 pKa = 4.16GGTYY231 pKa = 10.52DD232 pKa = 4.13FQSLEE237 pKa = 3.78IANPNVIFCAPGLKK251 pKa = 9.83EE252 pKa = 4.01QIEE255 pKa = 4.4SDD257 pKa = 3.49SRR259 pKa = 11.84FADD262 pKa = 4.78FQAVQQGNVVEE273 pKa = 4.19MDD275 pKa = 3.39EE276 pKa = 6.51SLMEE280 pKa = 3.95WQGRR284 pKa = 11.84TIVEE288 pKa = 3.86TAYY291 pKa = 9.98EE292 pKa = 3.88ISAAAFPQLLEE303 pKa = 4.35SSSSVSDD310 pKa = 3.3PTQDD314 pKa = 3.04IEE316 pKa = 4.72SQVSSALEE324 pKa = 3.67AQAQYY329 pKa = 8.07EE330 pKa = 4.21TLEE333 pKa = 4.23QGDD336 pKa = 3.76QGDD339 pKa = 4.19SVQALQEE346 pKa = 4.05RR347 pKa = 11.84LTQLGYY353 pKa = 9.14LTDD356 pKa = 5.22PYY358 pKa = 11.45DD359 pKa = 3.54GMYY362 pKa = 10.99GSTTADD368 pKa = 3.28CVSRR372 pKa = 11.84FQTANGLEE380 pKa = 4.16ATGIADD386 pKa = 4.16PATQAALFASGALRR400 pKa = 11.84EE401 pKa = 4.87DD402 pKa = 3.91GTPLPEE408 pKa = 4.11EE409 pKa = 4.43SGEE412 pKa = 4.28SSTSQSSSSASEE424 pKa = 4.22SSSSASEE431 pKa = 4.1SSASEE436 pKa = 4.16SSSQASSASSGSEE449 pKa = 3.7SSSQASASQQ458 pKa = 3.0
MM1 pKa = 7.02MRR3 pKa = 11.84IGSAIRR9 pKa = 11.84RR10 pKa = 11.84VLALALCVLLGCGAFAGCSQVDD32 pKa = 4.08DD33 pKa = 3.77IVSNVTAAGVFPVEE47 pKa = 4.4VNGVTISSRR56 pKa = 11.84PSRR59 pKa = 11.84VVVLSPSLADD69 pKa = 3.59VILALGCEE77 pKa = 4.17TQLAGASEE85 pKa = 4.96GCTQSALDD93 pKa = 3.84EE94 pKa = 4.44LEE96 pKa = 4.39KK97 pKa = 11.22VSADD101 pKa = 3.28SVEE104 pKa = 4.9AIQALQPDD112 pKa = 5.12LVLVDD117 pKa = 4.67PNSSGAQSALEE128 pKa = 3.99EE129 pKa = 4.39AGVTVLSVEE138 pKa = 4.24PATDD142 pKa = 3.21RR143 pKa = 11.84SDD145 pKa = 3.72FEE147 pKa = 4.45RR148 pKa = 11.84LYY150 pKa = 10.74SQVSSALLGGEE161 pKa = 3.87AGYY164 pKa = 11.03DD165 pKa = 3.35EE166 pKa = 6.1GIATAQDD173 pKa = 3.11IFITLDD179 pKa = 3.47NINRR183 pKa = 11.84IVPKK187 pKa = 9.9DD188 pKa = 3.78TITTACYY195 pKa = 10.13LYY197 pKa = 10.78DD198 pKa = 4.4LDD200 pKa = 4.67GSAVTGDD207 pKa = 3.71MFGSTIMSYY216 pKa = 11.07AGVTNVFEE224 pKa = 4.3SLEE227 pKa = 4.16GGTYY231 pKa = 10.52DD232 pKa = 4.13FQSLEE237 pKa = 3.78IANPNVIFCAPGLKK251 pKa = 9.83EE252 pKa = 4.01QIEE255 pKa = 4.4SDD257 pKa = 3.49SRR259 pKa = 11.84FADD262 pKa = 4.78FQAVQQGNVVEE273 pKa = 4.19MDD275 pKa = 3.39EE276 pKa = 6.51SLMEE280 pKa = 3.95WQGRR284 pKa = 11.84TIVEE288 pKa = 3.86TAYY291 pKa = 9.98EE292 pKa = 3.88ISAAAFPQLLEE303 pKa = 4.35SSSSVSDD310 pKa = 3.3PTQDD314 pKa = 3.04IEE316 pKa = 4.72SQVSSALEE324 pKa = 3.67AQAQYY329 pKa = 8.07EE330 pKa = 4.21TLEE333 pKa = 4.23QGDD336 pKa = 3.76QGDD339 pKa = 4.19SVQALQEE346 pKa = 4.05RR347 pKa = 11.84LTQLGYY353 pKa = 9.14LTDD356 pKa = 5.22PYY358 pKa = 11.45DD359 pKa = 3.54GMYY362 pKa = 10.99GSTTADD368 pKa = 3.28CVSRR372 pKa = 11.84FQTANGLEE380 pKa = 4.16ATGIADD386 pKa = 4.16PATQAALFASGALRR400 pKa = 11.84EE401 pKa = 4.87DD402 pKa = 3.91GTPLPEE408 pKa = 4.11EE409 pKa = 4.43SGEE412 pKa = 4.28SSTSQSSSSASEE424 pKa = 4.22SSSSASEE431 pKa = 4.1SSASEE436 pKa = 4.16SSSQASSASSGSEE449 pKa = 3.7SSSQASASQQ458 pKa = 3.0
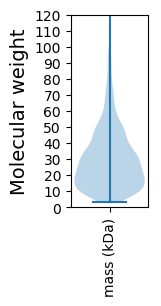
Molecular weight: 47.52 kDa
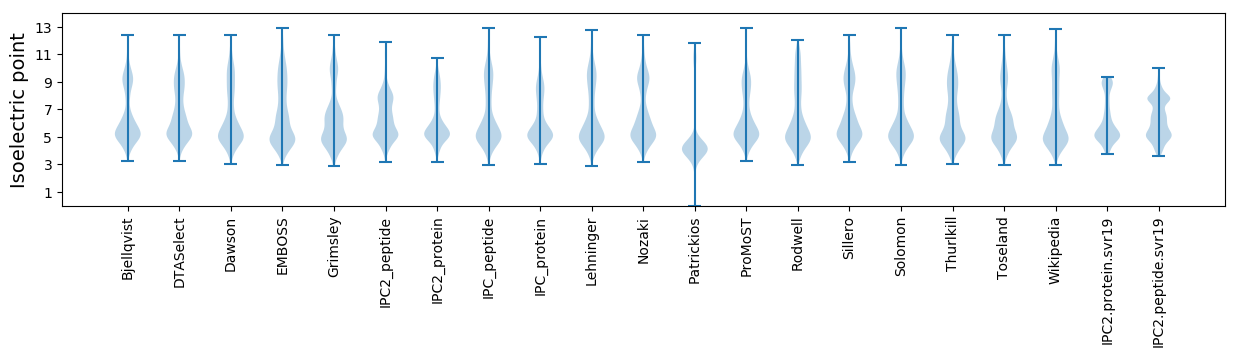
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R6YBB9|R6YBB9_9FIRM Oxidoreductase domain protein OS=Firmicutes bacterium CAG:94 OX=1262989 GN=BN815_02359 PE=4 SV=1
MM1 pKa = 7.8KK2 pKa = 10.08DD3 pKa = 3.33YY4 pKa = 10.99ILGVDD9 pKa = 4.64LGGTNIKK16 pKa = 9.97AAAYY20 pKa = 10.2RR21 pKa = 11.84LGSYY25 pKa = 10.03EE26 pKa = 3.9KK27 pKa = 10.61VGEE30 pKa = 4.22KK31 pKa = 10.22RR32 pKa = 11.84LPTQVEE38 pKa = 4.61GGWEE42 pKa = 4.1HH43 pKa = 6.08VLGRR47 pKa = 11.84VLAALEE53 pKa = 3.89EE54 pKa = 4.53LLRR57 pKa = 11.84HH58 pKa = 5.32TPRR61 pKa = 11.84EE62 pKa = 3.79RR63 pKa = 11.84VLCVGMGVPGLPGGGEE79 pKa = 4.24PCPGHH84 pKa = 7.07LPGGVRR90 pKa = 11.84NRR92 pKa = 11.84HH93 pKa = 5.35RR94 pKa = 11.84PAGGRR99 pKa = 11.84RR100 pKa = 11.84RR101 pKa = 11.84DD102 pKa = 3.4AGGCRR107 pKa = 11.84LRR109 pKa = 11.84RR110 pKa = 11.84PPHH113 pKa = 5.58GGGRR117 pKa = 11.84PDD119 pKa = 3.3VSRR122 pKa = 11.84KK123 pKa = 9.85KK124 pKa = 10.67SFPFF128 pKa = 4.04
MM1 pKa = 7.8KK2 pKa = 10.08DD3 pKa = 3.33YY4 pKa = 10.99ILGVDD9 pKa = 4.64LGGTNIKK16 pKa = 9.97AAAYY20 pKa = 10.2RR21 pKa = 11.84LGSYY25 pKa = 10.03EE26 pKa = 3.9KK27 pKa = 10.61VGEE30 pKa = 4.22KK31 pKa = 10.22RR32 pKa = 11.84LPTQVEE38 pKa = 4.61GGWEE42 pKa = 4.1HH43 pKa = 6.08VLGRR47 pKa = 11.84VLAALEE53 pKa = 3.89EE54 pKa = 4.53LLRR57 pKa = 11.84HH58 pKa = 5.32TPRR61 pKa = 11.84EE62 pKa = 3.79RR63 pKa = 11.84VLCVGMGVPGLPGGGEE79 pKa = 4.24PCPGHH84 pKa = 7.07LPGGVRR90 pKa = 11.84NRR92 pKa = 11.84HH93 pKa = 5.35RR94 pKa = 11.84PAGGRR99 pKa = 11.84RR100 pKa = 11.84RR101 pKa = 11.84DD102 pKa = 3.4AGGCRR107 pKa = 11.84LRR109 pKa = 11.84RR110 pKa = 11.84PPHH113 pKa = 5.58GGGRR117 pKa = 11.84PDD119 pKa = 3.3VSRR122 pKa = 11.84KK123 pKa = 9.85KK124 pKa = 10.67SFPFF128 pKa = 4.04
Molecular weight: 13.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
675675 |
29 |
1557 |
280.4 |
30.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.319 ± 0.06 | 1.62 ± 0.024 |
5.051 ± 0.039 | 7.162 ± 0.058 |
3.972 ± 0.033 | 8.262 ± 0.057 |
1.938 ± 0.028 | 4.975 ± 0.046 |
4.794 ± 0.045 | 10.376 ± 0.072 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.573 ± 0.024 | 3.114 ± 0.034 |
4.539 ± 0.036 | 4.233 ± 0.031 |
5.436 ± 0.052 | 5.496 ± 0.046 |
5.19 ± 0.043 | 7.378 ± 0.046 |
1.12 ± 0.019 | 3.446 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
