
Hubei sobemo-like virus 14
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.9
Get precalculated fractions of proteins
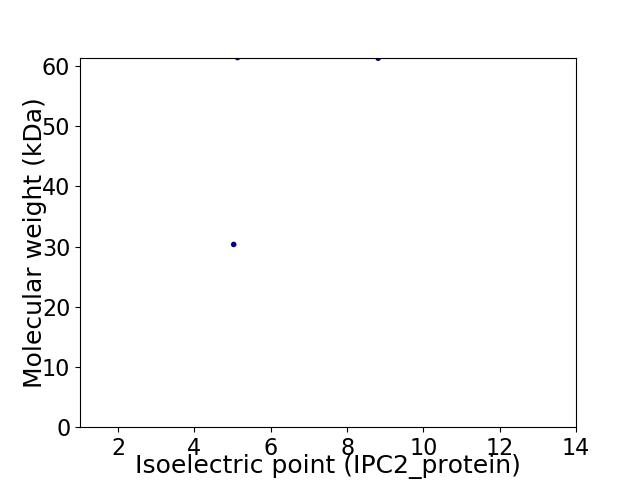
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEK1|A0A1L3KEK1_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 14 OX=1923199 PE=4 SV=1
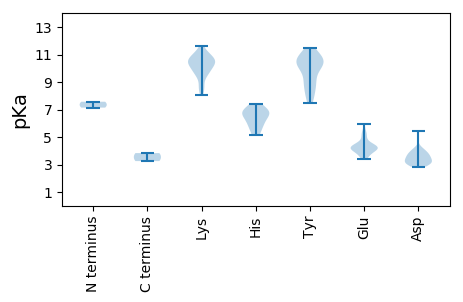
MM1 pKa = 7.57VDD3 pKa = 3.25TMVDD7 pKa = 2.87RR8 pKa = 11.84LLFQEE13 pKa = 4.01WTANVLKK20 pKa = 10.8KK21 pKa = 10.36HH22 pKa = 6.03MDD24 pKa = 3.31LPLAMGWAPISGGYY38 pKa = 8.91RR39 pKa = 11.84QLLRR43 pKa = 11.84RR44 pKa = 11.84FPGKK48 pKa = 10.55ALMADD53 pKa = 3.34KK54 pKa = 11.09SSWDD58 pKa = 3.08WTAQSWVFEE67 pKa = 4.12ALLAVLFEE75 pKa = 5.14LGPDD79 pKa = 3.35NDD81 pKa = 3.19CWRR84 pKa = 11.84RR85 pKa = 11.84VARR88 pKa = 11.84GRR90 pKa = 11.84FRR92 pKa = 11.84ALFYY96 pKa = 10.22EE97 pKa = 4.36AVMQFSDD104 pKa = 3.7GTLIQQDD111 pKa = 3.79YY112 pKa = 8.31TGVMKK117 pKa = 10.57SGCFLTLAGNSVAQLLLTSLAKK139 pKa = 10.44LRR141 pKa = 11.84LGWSDD146 pKa = 3.94SGEE149 pKa = 3.93FWALGDD155 pKa = 3.84DD156 pKa = 4.2TIEE159 pKa = 4.33DD160 pKa = 3.68VVPDD164 pKa = 3.05VEE166 pKa = 5.19RR167 pKa = 11.84YY168 pKa = 9.24VKK170 pKa = 10.12EE171 pKa = 4.05LQSCGCVIKK180 pKa = 10.12EE181 pKa = 3.92YY182 pKa = 10.75SVSDD186 pKa = 3.34VLEE189 pKa = 3.95FVGFVFHH196 pKa = 7.43RR197 pKa = 11.84DD198 pKa = 3.64SIPTPSYY205 pKa = 9.14VDD207 pKa = 2.85KK208 pKa = 11.07HH209 pKa = 5.54VFQIEE214 pKa = 4.56HH215 pKa = 6.84IDD217 pKa = 3.54DD218 pKa = 3.66AVYY221 pKa = 10.89EE222 pKa = 4.33EE223 pKa = 4.68TLQQYY228 pKa = 11.09LLLHH232 pKa = 6.65LNSPLKK238 pKa = 10.53PYY240 pKa = 10.63LDD242 pKa = 3.57RR243 pKa = 11.84LAEE246 pKa = 4.18RR247 pKa = 11.84EE248 pKa = 3.89PAGTFMSDD256 pKa = 3.07QKK258 pKa = 11.2LKK260 pKa = 10.78RR261 pKa = 11.84IWLGG265 pKa = 3.25
MM1 pKa = 7.57VDD3 pKa = 3.25TMVDD7 pKa = 2.87RR8 pKa = 11.84LLFQEE13 pKa = 4.01WTANVLKK20 pKa = 10.8KK21 pKa = 10.36HH22 pKa = 6.03MDD24 pKa = 3.31LPLAMGWAPISGGYY38 pKa = 8.91RR39 pKa = 11.84QLLRR43 pKa = 11.84RR44 pKa = 11.84FPGKK48 pKa = 10.55ALMADD53 pKa = 3.34KK54 pKa = 11.09SSWDD58 pKa = 3.08WTAQSWVFEE67 pKa = 4.12ALLAVLFEE75 pKa = 5.14LGPDD79 pKa = 3.35NDD81 pKa = 3.19CWRR84 pKa = 11.84RR85 pKa = 11.84VARR88 pKa = 11.84GRR90 pKa = 11.84FRR92 pKa = 11.84ALFYY96 pKa = 10.22EE97 pKa = 4.36AVMQFSDD104 pKa = 3.7GTLIQQDD111 pKa = 3.79YY112 pKa = 8.31TGVMKK117 pKa = 10.57SGCFLTLAGNSVAQLLLTSLAKK139 pKa = 10.44LRR141 pKa = 11.84LGWSDD146 pKa = 3.94SGEE149 pKa = 3.93FWALGDD155 pKa = 3.84DD156 pKa = 4.2TIEE159 pKa = 4.33DD160 pKa = 3.68VVPDD164 pKa = 3.05VEE166 pKa = 5.19RR167 pKa = 11.84YY168 pKa = 9.24VKK170 pKa = 10.12EE171 pKa = 4.05LQSCGCVIKK180 pKa = 10.12EE181 pKa = 3.92YY182 pKa = 10.75SVSDD186 pKa = 3.34VLEE189 pKa = 3.95FVGFVFHH196 pKa = 7.43RR197 pKa = 11.84DD198 pKa = 3.64SIPTPSYY205 pKa = 9.14VDD207 pKa = 2.85KK208 pKa = 11.07HH209 pKa = 5.54VFQIEE214 pKa = 4.56HH215 pKa = 6.84IDD217 pKa = 3.54DD218 pKa = 3.66AVYY221 pKa = 10.89EE222 pKa = 4.33EE223 pKa = 4.68TLQQYY228 pKa = 11.09LLLHH232 pKa = 6.65LNSPLKK238 pKa = 10.53PYY240 pKa = 10.63LDD242 pKa = 3.57RR243 pKa = 11.84LAEE246 pKa = 4.18RR247 pKa = 11.84EE248 pKa = 3.89PAGTFMSDD256 pKa = 3.07QKK258 pKa = 11.2LKK260 pKa = 10.78RR261 pKa = 11.84IWLGG265 pKa = 3.25
Molecular weight: 30.36 kDa
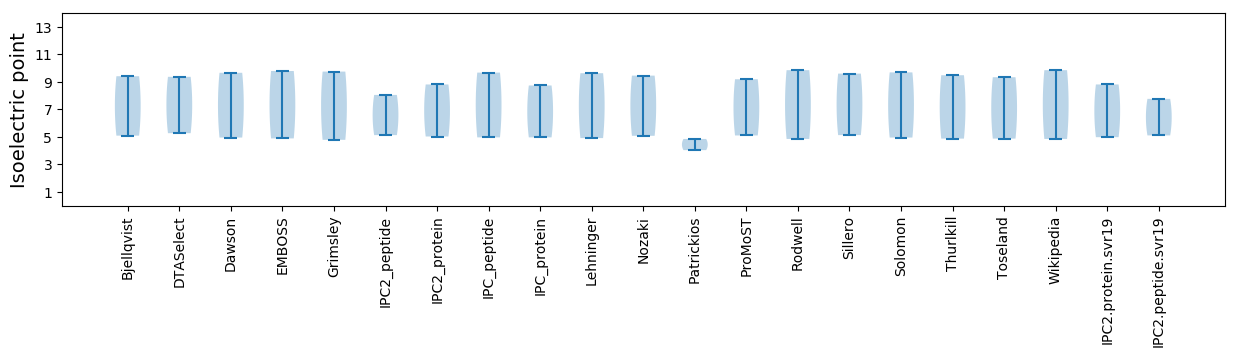
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEK1|A0A1L3KEK1_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 14 OX=1923199 PE=4 SV=1
MM1 pKa = 7.14SQTVMSIVDD10 pKa = 3.97RR11 pKa = 11.84LSEE14 pKa = 4.14PQWFRR19 pKa = 11.84NVEE22 pKa = 4.18SMSNIALPWVLTTLMAFYY40 pKa = 10.72LFVPPILGGLRR51 pKa = 11.84RR52 pKa = 11.84VARR55 pKa = 11.84NLIWLVVSVRR65 pKa = 11.84MACKK69 pKa = 10.15VFWRR73 pKa = 11.84NTRR76 pKa = 11.84DD77 pKa = 3.3SFLGQPRR84 pKa = 11.84YY85 pKa = 10.15NPEE88 pKa = 3.52STMPGSPFVPVSKK101 pKa = 10.55LPKK104 pKa = 8.04FQCMVLGNSRR114 pKa = 11.84AGADD118 pKa = 3.49PVQVGVAFRR127 pKa = 11.84FEE129 pKa = 4.24DD130 pKa = 3.98WIVTAWHH137 pKa = 5.89VVEE140 pKa = 5.43CCSEE144 pKa = 3.59MWLSVEE150 pKa = 4.38GEE152 pKa = 4.3TVPISKK158 pKa = 10.45SSFDD162 pKa = 3.72HH163 pKa = 6.94LEE165 pKa = 3.63GDD167 pKa = 3.01IAYY170 pKa = 10.14APFSKK175 pKa = 10.77DD176 pKa = 2.89LGRR179 pKa = 11.84LGLSKK184 pKa = 10.66PKK186 pKa = 10.07IRR188 pKa = 11.84TAGTGRR194 pKa = 11.84FDD196 pKa = 3.29EE197 pKa = 5.97AYY199 pKa = 9.12VQVAGGSPLSQTNGALKK216 pKa = 9.88PSRR219 pKa = 11.84SFGYY223 pKa = 10.07VRR225 pKa = 11.84YY226 pKa = 8.11TGSTKK231 pKa = 10.31PGFSGAPYY239 pKa = 7.51YY240 pKa = 10.38QHH242 pKa = 6.31VFVYY246 pKa = 10.3GMHH249 pKa = 6.93LGGGTEE255 pKa = 4.01NLGLDD260 pKa = 3.46ATYY263 pKa = 10.64LHH265 pKa = 6.88SMLLATRR272 pKa = 11.84EE273 pKa = 4.23STPDD277 pKa = 3.1ILDD280 pKa = 3.0KK281 pKa = 11.65VMFGRR286 pKa = 11.84QPIAVRR292 pKa = 11.84SFGNKK297 pKa = 8.61YY298 pKa = 7.7MAKK301 pKa = 9.51TGSGYY306 pKa = 10.85KK307 pKa = 10.28NIMYY311 pKa = 9.92EE312 pKa = 4.44DD313 pKa = 4.03YY314 pKa = 11.49VEE316 pKa = 4.91LSNQGRR322 pKa = 11.84SWADD326 pKa = 3.01MVEE329 pKa = 4.38DD330 pKa = 4.08YY331 pKa = 11.09EE332 pKa = 5.44GEE334 pKa = 4.21ANLMYY339 pKa = 10.61DD340 pKa = 4.21DD341 pKa = 5.43EE342 pKa = 5.05EE343 pKa = 4.47PTLQEE348 pKa = 4.04NFNRR352 pKa = 11.84AALRR356 pKa = 11.84AARR359 pKa = 11.84VAASEE364 pKa = 4.28TSSQLSVISGAVSSRR379 pKa = 11.84CMRR382 pKa = 11.84KK383 pKa = 8.43DD384 pKa = 3.3TEE386 pKa = 4.08IQEE389 pKa = 4.34LRR391 pKa = 11.84DD392 pKa = 3.54QISQLRR398 pKa = 11.84KK399 pKa = 9.77LLINYY404 pKa = 8.83HH405 pKa = 5.13EE406 pKa = 4.63QILVQGKK413 pKa = 9.39IPTQTTEE420 pKa = 3.69TSTSEE425 pKa = 4.46LKK427 pKa = 10.52KK428 pKa = 9.46QQEE431 pKa = 4.18NLRR434 pKa = 11.84QKK436 pKa = 11.09SSGQLSKK443 pKa = 8.9TQKK446 pKa = 9.73KK447 pKa = 9.73RR448 pKa = 11.84LRR450 pKa = 11.84QRR452 pKa = 11.84LSKK455 pKa = 10.78NITGEE460 pKa = 3.48PSMRR464 pKa = 11.84ILKK467 pKa = 8.57EE468 pKa = 3.39HH469 pKa = 6.54SGILTQDD476 pKa = 3.11QPPAYY481 pKa = 8.42VTYY484 pKa = 10.21KK485 pKa = 9.58GTVRR489 pKa = 11.84PLEE492 pKa = 4.13KK493 pKa = 10.64SLDD496 pKa = 3.79GMGSGTQDD504 pKa = 3.03LRR506 pKa = 11.84SWLSSSVKK514 pKa = 10.23YY515 pKa = 10.38SAEE518 pKa = 3.79SGALKK523 pKa = 10.63GEE525 pKa = 4.28KK526 pKa = 9.9LLSSTQSRR534 pKa = 11.84SSLSRR539 pKa = 11.84RR540 pKa = 11.84RR541 pKa = 11.84TSRR544 pKa = 11.84QKK546 pKa = 11.18LEE548 pKa = 3.83
MM1 pKa = 7.14SQTVMSIVDD10 pKa = 3.97RR11 pKa = 11.84LSEE14 pKa = 4.14PQWFRR19 pKa = 11.84NVEE22 pKa = 4.18SMSNIALPWVLTTLMAFYY40 pKa = 10.72LFVPPILGGLRR51 pKa = 11.84RR52 pKa = 11.84VARR55 pKa = 11.84NLIWLVVSVRR65 pKa = 11.84MACKK69 pKa = 10.15VFWRR73 pKa = 11.84NTRR76 pKa = 11.84DD77 pKa = 3.3SFLGQPRR84 pKa = 11.84YY85 pKa = 10.15NPEE88 pKa = 3.52STMPGSPFVPVSKK101 pKa = 10.55LPKK104 pKa = 8.04FQCMVLGNSRR114 pKa = 11.84AGADD118 pKa = 3.49PVQVGVAFRR127 pKa = 11.84FEE129 pKa = 4.24DD130 pKa = 3.98WIVTAWHH137 pKa = 5.89VVEE140 pKa = 5.43CCSEE144 pKa = 3.59MWLSVEE150 pKa = 4.38GEE152 pKa = 4.3TVPISKK158 pKa = 10.45SSFDD162 pKa = 3.72HH163 pKa = 6.94LEE165 pKa = 3.63GDD167 pKa = 3.01IAYY170 pKa = 10.14APFSKK175 pKa = 10.77DD176 pKa = 2.89LGRR179 pKa = 11.84LGLSKK184 pKa = 10.66PKK186 pKa = 10.07IRR188 pKa = 11.84TAGTGRR194 pKa = 11.84FDD196 pKa = 3.29EE197 pKa = 5.97AYY199 pKa = 9.12VQVAGGSPLSQTNGALKK216 pKa = 9.88PSRR219 pKa = 11.84SFGYY223 pKa = 10.07VRR225 pKa = 11.84YY226 pKa = 8.11TGSTKK231 pKa = 10.31PGFSGAPYY239 pKa = 7.51YY240 pKa = 10.38QHH242 pKa = 6.31VFVYY246 pKa = 10.3GMHH249 pKa = 6.93LGGGTEE255 pKa = 4.01NLGLDD260 pKa = 3.46ATYY263 pKa = 10.64LHH265 pKa = 6.88SMLLATRR272 pKa = 11.84EE273 pKa = 4.23STPDD277 pKa = 3.1ILDD280 pKa = 3.0KK281 pKa = 11.65VMFGRR286 pKa = 11.84QPIAVRR292 pKa = 11.84SFGNKK297 pKa = 8.61YY298 pKa = 7.7MAKK301 pKa = 9.51TGSGYY306 pKa = 10.85KK307 pKa = 10.28NIMYY311 pKa = 9.92EE312 pKa = 4.44DD313 pKa = 4.03YY314 pKa = 11.49VEE316 pKa = 4.91LSNQGRR322 pKa = 11.84SWADD326 pKa = 3.01MVEE329 pKa = 4.38DD330 pKa = 4.08YY331 pKa = 11.09EE332 pKa = 5.44GEE334 pKa = 4.21ANLMYY339 pKa = 10.61DD340 pKa = 4.21DD341 pKa = 5.43EE342 pKa = 5.05EE343 pKa = 4.47PTLQEE348 pKa = 4.04NFNRR352 pKa = 11.84AALRR356 pKa = 11.84AARR359 pKa = 11.84VAASEE364 pKa = 4.28TSSQLSVISGAVSSRR379 pKa = 11.84CMRR382 pKa = 11.84KK383 pKa = 8.43DD384 pKa = 3.3TEE386 pKa = 4.08IQEE389 pKa = 4.34LRR391 pKa = 11.84DD392 pKa = 3.54QISQLRR398 pKa = 11.84KK399 pKa = 9.77LLINYY404 pKa = 8.83HH405 pKa = 5.13EE406 pKa = 4.63QILVQGKK413 pKa = 9.39IPTQTTEE420 pKa = 3.69TSTSEE425 pKa = 4.46LKK427 pKa = 10.52KK428 pKa = 9.46QQEE431 pKa = 4.18NLRR434 pKa = 11.84QKK436 pKa = 11.09SSGQLSKK443 pKa = 8.9TQKK446 pKa = 9.73KK447 pKa = 9.73RR448 pKa = 11.84LRR450 pKa = 11.84QRR452 pKa = 11.84LSKK455 pKa = 10.78NITGEE460 pKa = 3.48PSMRR464 pKa = 11.84ILKK467 pKa = 8.57EE468 pKa = 3.39HH469 pKa = 6.54SGILTQDD476 pKa = 3.11QPPAYY481 pKa = 8.42VTYY484 pKa = 10.21KK485 pKa = 9.58GTVRR489 pKa = 11.84PLEE492 pKa = 4.13KK493 pKa = 10.64SLDD496 pKa = 3.79GMGSGTQDD504 pKa = 3.03LRR506 pKa = 11.84SWLSSSVKK514 pKa = 10.23YY515 pKa = 10.38SAEE518 pKa = 3.79SGALKK523 pKa = 10.63GEE525 pKa = 4.28KK526 pKa = 9.9LLSSTQSRR534 pKa = 11.84SSLSRR539 pKa = 11.84RR540 pKa = 11.84RR541 pKa = 11.84TSRR544 pKa = 11.84QKK546 pKa = 11.18LEE548 pKa = 3.83
Molecular weight: 61.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
813 |
265 |
548 |
406.5 |
45.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.027 ± 0.415 | 1.107 ± 0.218 |
5.166 ± 1.497 | 5.904 ± 0.132 |
3.936 ± 0.731 | 7.257 ± 0.457 |
1.476 ± 0.223 | 3.444 ± 0.231 |
5.166 ± 0.346 | 10.701 ± 1.36 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.198 ± 0.097 | 2.583 ± 0.583 |
4.428 ± 0.355 | 5.043 ± 0.279 |
6.642 ± 0.533 | 9.594 ± 1.725 |
5.412 ± 0.684 | 7.134 ± 0.429 |
2.214 ± 0.642 | 3.567 ± 0.093 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
