
Actinoplanes teichomyceticus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micromonosporales; Micromonosporaceae; Actinoplanes
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
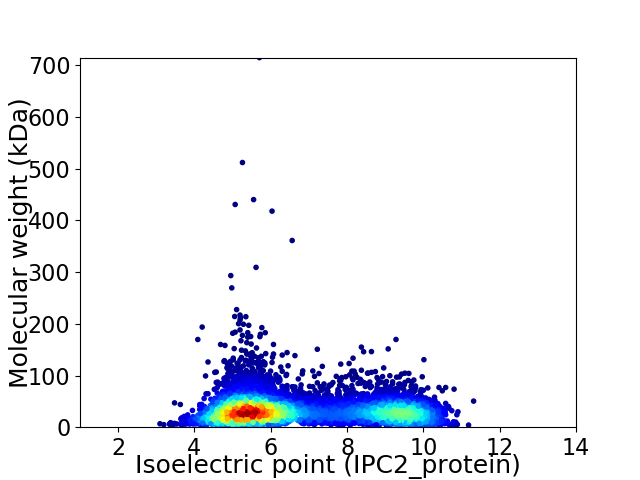
Virtual 2D-PAGE plot for 7203 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A561WJN4|A0A561WJN4_ACTTI Glycosyl transferase family 2 OS=Actinoplanes teichomyceticus OX=1867 GN=FHX34_102652 PE=4 SV=1
MM1 pKa = 7.18TFEE4 pKa = 4.44NVGPASDD11 pKa = 3.44IAKK14 pKa = 8.85GTVLQVEE21 pKa = 4.16IDD23 pKa = 3.7GVEE26 pKa = 4.0IAVVHH31 pKa = 6.9ADD33 pKa = 3.4DD34 pKa = 4.57DD35 pKa = 3.83NYY37 pKa = 11.16YY38 pKa = 10.96AVRR41 pKa = 11.84DD42 pKa = 4.09EE43 pKa = 4.7CSHH46 pKa = 6.77ASVALSEE53 pKa = 4.65GEE55 pKa = 3.95VDD57 pKa = 4.97GCTLEE62 pKa = 4.28CWLHH66 pKa = 6.03GSRR69 pKa = 11.84FDD71 pKa = 4.61LRR73 pKa = 11.84TGEE76 pKa = 4.11PSGPPAIDD84 pKa = 3.3PVAVYY89 pKa = 9.48PVEE92 pKa = 4.29IRR94 pKa = 11.84DD95 pKa = 3.22GDD97 pKa = 3.87IYY99 pKa = 11.59VSTTPSNGVEE109 pKa = 4.05PP110 pKa = 4.39
MM1 pKa = 7.18TFEE4 pKa = 4.44NVGPASDD11 pKa = 3.44IAKK14 pKa = 8.85GTVLQVEE21 pKa = 4.16IDD23 pKa = 3.7GVEE26 pKa = 4.0IAVVHH31 pKa = 6.9ADD33 pKa = 3.4DD34 pKa = 4.57DD35 pKa = 3.83NYY37 pKa = 11.16YY38 pKa = 10.96AVRR41 pKa = 11.84DD42 pKa = 4.09EE43 pKa = 4.7CSHH46 pKa = 6.77ASVALSEE53 pKa = 4.65GEE55 pKa = 3.95VDD57 pKa = 4.97GCTLEE62 pKa = 4.28CWLHH66 pKa = 6.03GSRR69 pKa = 11.84FDD71 pKa = 4.61LRR73 pKa = 11.84TGEE76 pKa = 4.11PSGPPAIDD84 pKa = 3.3PVAVYY89 pKa = 9.48PVEE92 pKa = 4.29IRR94 pKa = 11.84DD95 pKa = 3.22GDD97 pKa = 3.87IYY99 pKa = 11.59VSTTPSNGVEE109 pKa = 4.05PP110 pKa = 4.39
Molecular weight: 11.71 kDa
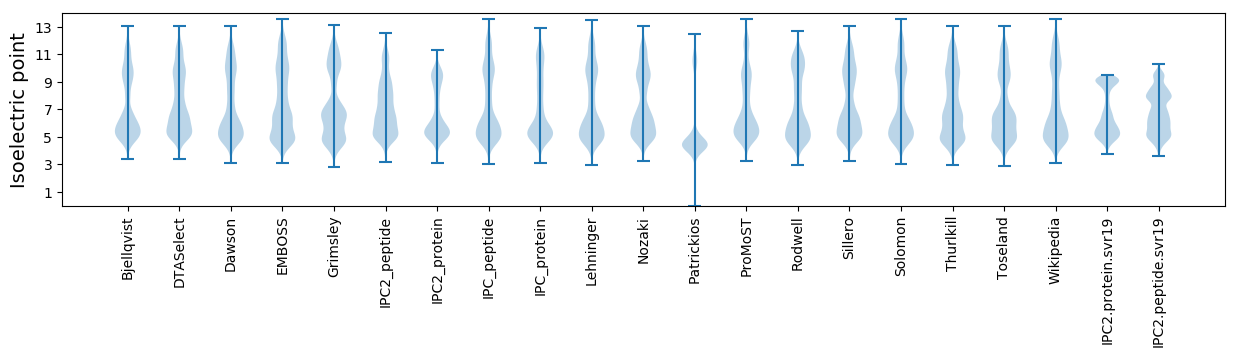
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A561WLD8|A0A561WLD8_ACTTI DNA-binding SARP family transcriptional activator OS=Actinoplanes teichomyceticus OX=1867 GN=FHX34_1021242 PE=3 SV=1
MM1 pKa = 7.69ASHH4 pKa = 7.41PPALPPGEE12 pKa = 4.15RR13 pKa = 11.84PPRR16 pKa = 11.84HH17 pKa = 5.82SPAPRR22 pKa = 11.84SPHH25 pKa = 6.06HH26 pKa = 7.0PSRR29 pKa = 11.84PPWTRR34 pKa = 11.84AEE36 pKa = 4.08PPARR40 pKa = 11.84TRR42 pKa = 11.84RR43 pKa = 11.84PRR45 pKa = 11.84RR46 pKa = 11.84PRR48 pKa = 11.84PPRR51 pKa = 11.84APAARR56 pKa = 11.84RR57 pKa = 11.84RR58 pKa = 11.84HH59 pKa = 5.56RR60 pKa = 11.84SRR62 pKa = 11.84RR63 pKa = 11.84SPGPPPRR70 pKa = 11.84PPVPHH75 pKa = 6.39RR76 pKa = 11.84PSRR79 pKa = 11.84RR80 pKa = 11.84RR81 pKa = 11.84RR82 pKa = 11.84RR83 pKa = 11.84RR84 pKa = 11.84RR85 pKa = 11.84RR86 pKa = 11.84SPRR89 pKa = 11.84SPQPPRR95 pKa = 11.84RR96 pKa = 11.84TSPRR100 pKa = 11.84RR101 pKa = 11.84RR102 pKa = 11.84RR103 pKa = 11.84SRR105 pKa = 11.84ATGASRR111 pKa = 11.84LPSPARR117 pKa = 11.84RR118 pKa = 11.84RR119 pKa = 11.84AQVPRR124 pKa = 11.84GPRR127 pKa = 11.84PRR129 pKa = 11.84SPDD132 pKa = 3.04RR133 pKa = 11.84AKK135 pKa = 10.84ARR137 pKa = 11.84PRR139 pKa = 11.84PAAPCGAGPAGRR151 pKa = 11.84HH152 pKa = 4.16RR153 pKa = 11.84RR154 pKa = 11.84RR155 pKa = 11.84LRR157 pKa = 11.84VLRR160 pKa = 11.84RR161 pKa = 11.84PRR163 pKa = 11.84RR164 pKa = 11.84RR165 pKa = 11.84RR166 pKa = 11.84LRR168 pKa = 11.84VLRR171 pKa = 11.84RR172 pKa = 11.84PRR174 pKa = 11.84RR175 pKa = 11.84HH176 pKa = 5.73RR177 pKa = 11.84PGVLGRR183 pKa = 11.84PRR185 pKa = 11.84RR186 pKa = 11.84RR187 pKa = 11.84RR188 pKa = 11.84VGLSPPGLPSPSHH201 pKa = 5.95GSRR204 pKa = 11.84RR205 pKa = 11.84RR206 pKa = 11.84RR207 pKa = 11.84AVRR210 pKa = 11.84IPRR213 pKa = 11.84MPHH216 pKa = 5.16AAARR220 pKa = 11.84MPHH223 pKa = 6.2AAARR227 pKa = 11.84MPHH230 pKa = 6.2AAARR234 pKa = 11.84MPHH237 pKa = 6.2AAARR241 pKa = 11.84MPHH244 pKa = 6.2AAARR248 pKa = 11.84MPHH251 pKa = 6.2AAARR255 pKa = 11.84MPHH258 pKa = 6.2AAARR262 pKa = 11.84MPHH265 pKa = 6.46AAACGRR271 pKa = 11.84RR272 pKa = 11.84PRR274 pKa = 11.84RR275 pKa = 11.84KK276 pKa = 8.91PPQVRR281 pKa = 11.84PLPRR285 pKa = 11.84GRR287 pKa = 11.84NPGSRR292 pKa = 11.84ALSRR296 pKa = 11.84QARR299 pKa = 11.84PGPLRR304 pKa = 11.84GLRR307 pKa = 11.84RR308 pKa = 11.84RR309 pKa = 11.84RR310 pKa = 11.84SRR312 pKa = 11.84PSRR315 pKa = 11.84AAQPGQPSRR324 pKa = 11.84QPGRR328 pKa = 11.84LRR330 pKa = 11.84LLSRR334 pKa = 11.84QPGRR338 pKa = 11.84QRR340 pKa = 11.84RR341 pKa = 11.84GPGRR345 pKa = 11.84RR346 pKa = 11.84SHH348 pKa = 6.91RR349 pKa = 11.84SGRR352 pKa = 11.84LRR354 pKa = 11.84RR355 pKa = 11.84EE356 pKa = 3.77PGRR359 pKa = 11.84VRR361 pKa = 11.84RR362 pKa = 11.84QRR364 pKa = 11.84QRR366 pKa = 11.84RR367 pKa = 11.84GRR369 pKa = 11.84RR370 pKa = 11.84SPLRR374 pKa = 11.84SQARR378 pKa = 11.84RR379 pKa = 11.84TCRR382 pKa = 11.84GRR384 pKa = 11.84PWRR387 pKa = 11.84CRR389 pKa = 11.84PRR391 pKa = 11.84WLPAGRR397 pKa = 11.84RR398 pKa = 11.84RR399 pKa = 11.84GWTRR403 pKa = 11.84VQPSPPIRR411 pKa = 11.84PKK413 pKa = 9.95SAARR417 pKa = 11.84GAVLPGPGAGPVAEE431 pKa = 4.27PFGARR436 pKa = 11.84TALPAPSPP444 pKa = 3.76
MM1 pKa = 7.69ASHH4 pKa = 7.41PPALPPGEE12 pKa = 4.15RR13 pKa = 11.84PPRR16 pKa = 11.84HH17 pKa = 5.82SPAPRR22 pKa = 11.84SPHH25 pKa = 6.06HH26 pKa = 7.0PSRR29 pKa = 11.84PPWTRR34 pKa = 11.84AEE36 pKa = 4.08PPARR40 pKa = 11.84TRR42 pKa = 11.84RR43 pKa = 11.84PRR45 pKa = 11.84RR46 pKa = 11.84PRR48 pKa = 11.84PPRR51 pKa = 11.84APAARR56 pKa = 11.84RR57 pKa = 11.84RR58 pKa = 11.84HH59 pKa = 5.56RR60 pKa = 11.84SRR62 pKa = 11.84RR63 pKa = 11.84SPGPPPRR70 pKa = 11.84PPVPHH75 pKa = 6.39RR76 pKa = 11.84PSRR79 pKa = 11.84RR80 pKa = 11.84RR81 pKa = 11.84RR82 pKa = 11.84RR83 pKa = 11.84RR84 pKa = 11.84RR85 pKa = 11.84RR86 pKa = 11.84SPRR89 pKa = 11.84SPQPPRR95 pKa = 11.84RR96 pKa = 11.84TSPRR100 pKa = 11.84RR101 pKa = 11.84RR102 pKa = 11.84RR103 pKa = 11.84SRR105 pKa = 11.84ATGASRR111 pKa = 11.84LPSPARR117 pKa = 11.84RR118 pKa = 11.84RR119 pKa = 11.84AQVPRR124 pKa = 11.84GPRR127 pKa = 11.84PRR129 pKa = 11.84SPDD132 pKa = 3.04RR133 pKa = 11.84AKK135 pKa = 10.84ARR137 pKa = 11.84PRR139 pKa = 11.84PAAPCGAGPAGRR151 pKa = 11.84HH152 pKa = 4.16RR153 pKa = 11.84RR154 pKa = 11.84RR155 pKa = 11.84LRR157 pKa = 11.84VLRR160 pKa = 11.84RR161 pKa = 11.84PRR163 pKa = 11.84RR164 pKa = 11.84RR165 pKa = 11.84RR166 pKa = 11.84LRR168 pKa = 11.84VLRR171 pKa = 11.84RR172 pKa = 11.84PRR174 pKa = 11.84RR175 pKa = 11.84HH176 pKa = 5.73RR177 pKa = 11.84PGVLGRR183 pKa = 11.84PRR185 pKa = 11.84RR186 pKa = 11.84RR187 pKa = 11.84RR188 pKa = 11.84VGLSPPGLPSPSHH201 pKa = 5.95GSRR204 pKa = 11.84RR205 pKa = 11.84RR206 pKa = 11.84RR207 pKa = 11.84AVRR210 pKa = 11.84IPRR213 pKa = 11.84MPHH216 pKa = 5.16AAARR220 pKa = 11.84MPHH223 pKa = 6.2AAARR227 pKa = 11.84MPHH230 pKa = 6.2AAARR234 pKa = 11.84MPHH237 pKa = 6.2AAARR241 pKa = 11.84MPHH244 pKa = 6.2AAARR248 pKa = 11.84MPHH251 pKa = 6.2AAARR255 pKa = 11.84MPHH258 pKa = 6.2AAARR262 pKa = 11.84MPHH265 pKa = 6.46AAACGRR271 pKa = 11.84RR272 pKa = 11.84PRR274 pKa = 11.84RR275 pKa = 11.84KK276 pKa = 8.91PPQVRR281 pKa = 11.84PLPRR285 pKa = 11.84GRR287 pKa = 11.84NPGSRR292 pKa = 11.84ALSRR296 pKa = 11.84QARR299 pKa = 11.84PGPLRR304 pKa = 11.84GLRR307 pKa = 11.84RR308 pKa = 11.84RR309 pKa = 11.84RR310 pKa = 11.84SRR312 pKa = 11.84PSRR315 pKa = 11.84AAQPGQPSRR324 pKa = 11.84QPGRR328 pKa = 11.84LRR330 pKa = 11.84LLSRR334 pKa = 11.84QPGRR338 pKa = 11.84QRR340 pKa = 11.84RR341 pKa = 11.84GPGRR345 pKa = 11.84RR346 pKa = 11.84SHH348 pKa = 6.91RR349 pKa = 11.84SGRR352 pKa = 11.84LRR354 pKa = 11.84RR355 pKa = 11.84EE356 pKa = 3.77PGRR359 pKa = 11.84VRR361 pKa = 11.84RR362 pKa = 11.84QRR364 pKa = 11.84QRR366 pKa = 11.84RR367 pKa = 11.84GRR369 pKa = 11.84RR370 pKa = 11.84SPLRR374 pKa = 11.84SQARR378 pKa = 11.84RR379 pKa = 11.84TCRR382 pKa = 11.84GRR384 pKa = 11.84PWRR387 pKa = 11.84CRR389 pKa = 11.84PRR391 pKa = 11.84WLPAGRR397 pKa = 11.84RR398 pKa = 11.84RR399 pKa = 11.84GWTRR403 pKa = 11.84VQPSPPIRR411 pKa = 11.84PKK413 pKa = 9.95SAARR417 pKa = 11.84GAVLPGPGAGPVAEE431 pKa = 4.27PFGARR436 pKa = 11.84TALPAPSPP444 pKa = 3.76
Molecular weight: 50.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2375611 |
26 |
6666 |
329.8 |
35.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.587 ± 0.045 | 0.757 ± 0.007 |
5.9 ± 0.023 | 4.906 ± 0.028 |
2.649 ± 0.016 | 9.475 ± 0.033 |
2.193 ± 0.014 | 3.357 ± 0.018 |
1.644 ± 0.022 | 10.236 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.67 ± 0.012 | 1.775 ± 0.02 |
6.433 ± 0.034 | 2.712 ± 0.014 |
8.588 ± 0.034 | 4.775 ± 0.02 |
6.136 ± 0.034 | 8.593 ± 0.029 |
1.559 ± 0.014 | 2.054 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
