
Capybara microvirus Cap1_SP_94
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.38
Get precalculated fractions of proteins
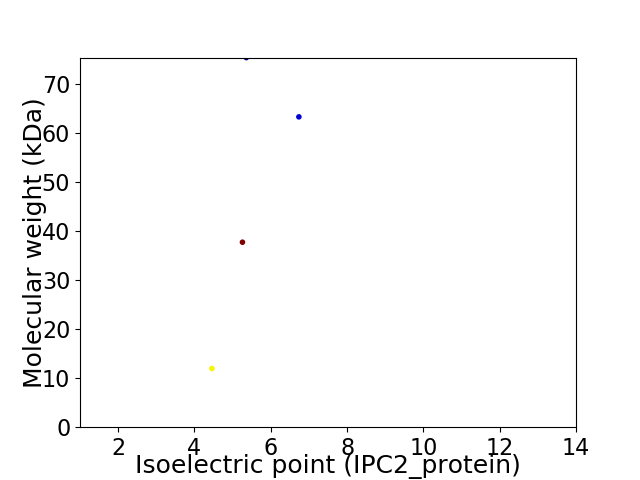
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
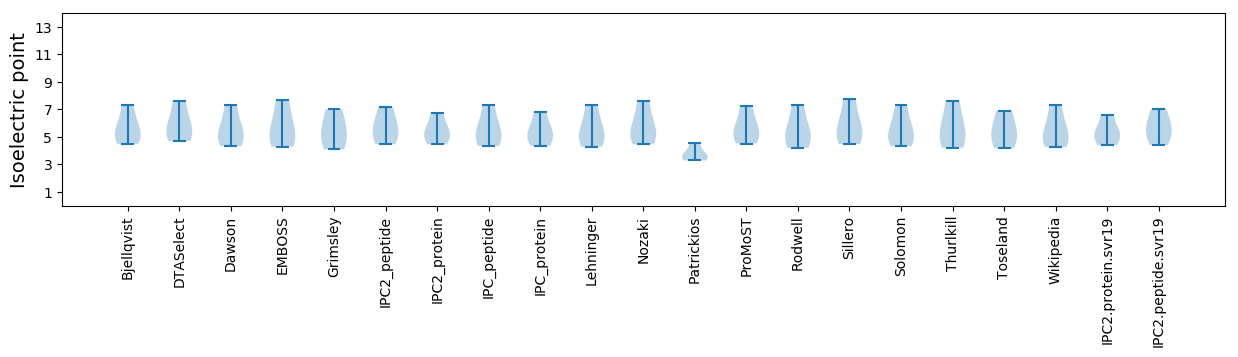
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W7N9|A0A4P8W7N9_9VIRU Uncharacterized protein OS=Capybara microvirus Cap1_SP_94 OX=2584800 PE=4 SV=1
MM1 pKa = 7.49FFPFTSYY8 pKa = 11.27SPDD11 pKa = 3.27LSGSEE16 pKa = 4.01VLEE19 pKa = 4.07RR20 pKa = 11.84KK21 pKa = 9.69YY22 pKa = 11.09DD23 pKa = 3.9LQTMIDD29 pKa = 4.11PNNSTLVLHH38 pKa = 7.0RR39 pKa = 11.84GLLTDD44 pKa = 3.65VPISFNSQYY53 pKa = 10.85LPKK56 pKa = 10.62PEE58 pKa = 4.34DD59 pKa = 3.38TDD61 pKa = 4.9LAKK64 pKa = 10.4QLQNGSPLEE73 pKa = 4.7DD74 pKa = 3.65YY75 pKa = 10.99SGSRR79 pKa = 11.84PFNGAAHH86 pKa = 7.01HH87 pKa = 6.79IDD89 pKa = 3.73DD90 pKa = 4.37VSANTYY96 pKa = 9.08MDD98 pKa = 4.31EE99 pKa = 4.87LSKK102 pKa = 9.74TLQTSS107 pKa = 3.39
MM1 pKa = 7.49FFPFTSYY8 pKa = 11.27SPDD11 pKa = 3.27LSGSEE16 pKa = 4.01VLEE19 pKa = 4.07RR20 pKa = 11.84KK21 pKa = 9.69YY22 pKa = 11.09DD23 pKa = 3.9LQTMIDD29 pKa = 4.11PNNSTLVLHH38 pKa = 7.0RR39 pKa = 11.84GLLTDD44 pKa = 3.65VPISFNSQYY53 pKa = 10.85LPKK56 pKa = 10.62PEE58 pKa = 4.34DD59 pKa = 3.38TDD61 pKa = 4.9LAKK64 pKa = 10.4QLQNGSPLEE73 pKa = 4.7DD74 pKa = 3.65YY75 pKa = 10.99SGSRR79 pKa = 11.84PFNGAAHH86 pKa = 7.01HH87 pKa = 6.79IDD89 pKa = 3.73DD90 pKa = 4.37VSANTYY96 pKa = 9.08MDD98 pKa = 4.31EE99 pKa = 4.87LSKK102 pKa = 9.74TLQTSS107 pKa = 3.39
Molecular weight: 11.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W544|A0A4P8W544_9VIRU Uncharacterized protein OS=Capybara microvirus Cap1_SP_94 OX=2584800 PE=4 SV=1
MM1 pKa = 7.73SICQNFKK8 pKa = 10.7HH9 pKa = 5.95IASRR13 pKa = 11.84SKK15 pKa = 10.69RR16 pKa = 11.84LRR18 pKa = 11.84ANLDD22 pKa = 3.01ALYY25 pKa = 11.27NKK27 pKa = 9.32VSCNKK32 pKa = 9.66CDD34 pKa = 3.08SCLAQNRR41 pKa = 11.84NDD43 pKa = 3.0WYY45 pKa = 10.1VRR47 pKa = 11.84ARR49 pKa = 11.84FEE51 pKa = 4.11MMQTLGLDD59 pKa = 3.45YY60 pKa = 10.82YY61 pKa = 10.73NNVVADD67 pKa = 3.97PGFILFPCLTYY78 pKa = 10.48IPTNRR83 pKa = 11.84PTLEE87 pKa = 4.29LLDD90 pKa = 5.37DD91 pKa = 4.32NSLPYY96 pKa = 10.41YY97 pKa = 10.66FEE99 pKa = 4.49GFSYY103 pKa = 10.59SHH105 pKa = 7.06IKK107 pKa = 10.34RR108 pKa = 11.84FLVEE112 pKa = 4.33LADD115 pKa = 3.82KK116 pKa = 9.98FQNRR120 pKa = 11.84YY121 pKa = 7.61HH122 pKa = 6.84TGTSCGVRR130 pKa = 11.84YY131 pKa = 8.46MVASEE136 pKa = 3.77YY137 pKa = 11.07GYY139 pKa = 11.27AEE141 pKa = 4.78EE142 pKa = 4.25YY143 pKa = 10.98LDD145 pKa = 5.34DD146 pKa = 5.04FGHH149 pKa = 6.37SRR151 pKa = 11.84IGQAAPHH158 pKa = 4.95YY159 pKa = 9.69HH160 pKa = 5.85VLLFFPSVAYY170 pKa = 10.23NDD172 pKa = 3.36KK173 pKa = 10.97DD174 pKa = 3.38YY175 pKa = 10.74WADD178 pKa = 3.39VVEE181 pKa = 4.65SLWSVTHH188 pKa = 4.55RR189 pKa = 11.84HH190 pKa = 5.57GRR192 pKa = 11.84VGWSKK197 pKa = 10.69KK198 pKa = 9.07HH199 pKa = 5.4GKK201 pKa = 9.44FVTSPLAIKK210 pKa = 9.16YY211 pKa = 5.33TTQYY215 pKa = 10.16CHH217 pKa = 6.53KK218 pKa = 9.46QHH220 pKa = 6.74SYY222 pKa = 7.52YY223 pKa = 10.24QRR225 pKa = 11.84PEE227 pKa = 3.64IEE229 pKa = 3.49KK230 pKa = 9.78WLYY233 pKa = 10.58QDD235 pKa = 3.29ITKK238 pKa = 8.14DD239 pKa = 3.52TPFHH243 pKa = 5.72VQHH246 pKa = 6.29EE247 pKa = 4.63LIKK250 pKa = 10.47EE251 pKa = 4.07RR252 pKa = 11.84EE253 pKa = 4.11KK254 pKa = 10.86QIHH257 pKa = 7.4DD258 pKa = 3.31ILPFHH263 pKa = 7.23RR264 pKa = 11.84EE265 pKa = 3.69SLHH268 pKa = 7.05FGEE271 pKa = 5.38CAVDD275 pKa = 4.0FLKK278 pKa = 11.04SLSSEE283 pKa = 3.9EE284 pKa = 4.15RR285 pKa = 11.84IKK287 pKa = 11.09LLSNGITFPDD297 pKa = 4.07DD298 pKa = 3.3FAADD302 pKa = 3.67GTLNHH307 pKa = 6.88YY308 pKa = 10.15ILPKK312 pKa = 10.23YY313 pKa = 10.33IYY315 pKa = 9.78DD316 pKa = 3.4KK317 pKa = 11.09LYY319 pKa = 10.58FVIEE323 pKa = 4.16NKK325 pKa = 10.42SVIEE329 pKa = 4.35DD330 pKa = 3.43GKK332 pKa = 11.02SVVKK336 pKa = 10.07PFVRR340 pKa = 11.84YY341 pKa = 9.92GSDD344 pKa = 3.27YY345 pKa = 11.65ALADD349 pKa = 3.35YY350 pKa = 7.5RR351 pKa = 11.84TLFRR355 pKa = 11.84YY356 pKa = 9.22NFNKK360 pKa = 10.11SRR362 pKa = 11.84FDD364 pKa = 3.72LEE366 pKa = 5.28RR367 pKa = 11.84ITSNLFLSQLDD378 pKa = 3.56KK379 pKa = 11.47EE380 pKa = 4.95LIEE383 pKa = 4.45KK384 pKa = 10.13LNKK387 pKa = 9.16RR388 pKa = 11.84DD389 pKa = 3.66YY390 pKa = 10.6VLQYY394 pKa = 10.17NYY396 pKa = 11.17ADD398 pKa = 3.56FRR400 pKa = 11.84HH401 pKa = 6.53FIDD404 pKa = 5.65DD405 pKa = 3.14IKK407 pKa = 11.32SRR409 pKa = 11.84YY410 pKa = 7.3TFDD413 pKa = 5.63RR414 pKa = 11.84ILSYY418 pKa = 10.96KK419 pKa = 10.3HH420 pKa = 6.13IVHH423 pKa = 6.71HH424 pKa = 6.63LFAFDD429 pKa = 3.95PLSYY433 pKa = 11.1SDD435 pKa = 4.92DD436 pKa = 3.57DD437 pKa = 4.52LYY439 pKa = 11.96NFVEE443 pKa = 5.13DD444 pKa = 3.44ISVNKK449 pKa = 10.13ILTKK453 pKa = 10.43RR454 pKa = 11.84FSSLDD459 pKa = 3.33KK460 pKa = 11.23YY461 pKa = 10.82EE462 pKa = 3.92LSIYY466 pKa = 10.29QKK468 pKa = 10.88LFSSYY473 pKa = 10.17NSPDD477 pKa = 2.91ISYY480 pKa = 10.9LDD482 pKa = 4.28GYY484 pKa = 10.75SCVRR488 pKa = 11.84NDD490 pKa = 3.89EE491 pKa = 4.17LDD493 pKa = 3.54EE494 pKa = 4.34FLSIIDD500 pKa = 4.24TIKK503 pKa = 10.62LHH505 pKa = 7.11ASVHH509 pKa = 5.53RR510 pKa = 11.84KK511 pKa = 8.77KK512 pKa = 10.8AYY514 pKa = 10.36DD515 pKa = 3.66LVCLQKK521 pKa = 10.3QAHH524 pKa = 7.17KK525 pKa = 10.57YY526 pKa = 9.84RR527 pKa = 11.84NLSKK531 pKa = 11.2YY532 pKa = 10.22ILL534 pKa = 3.91
MM1 pKa = 7.73SICQNFKK8 pKa = 10.7HH9 pKa = 5.95IASRR13 pKa = 11.84SKK15 pKa = 10.69RR16 pKa = 11.84LRR18 pKa = 11.84ANLDD22 pKa = 3.01ALYY25 pKa = 11.27NKK27 pKa = 9.32VSCNKK32 pKa = 9.66CDD34 pKa = 3.08SCLAQNRR41 pKa = 11.84NDD43 pKa = 3.0WYY45 pKa = 10.1VRR47 pKa = 11.84ARR49 pKa = 11.84FEE51 pKa = 4.11MMQTLGLDD59 pKa = 3.45YY60 pKa = 10.82YY61 pKa = 10.73NNVVADD67 pKa = 3.97PGFILFPCLTYY78 pKa = 10.48IPTNRR83 pKa = 11.84PTLEE87 pKa = 4.29LLDD90 pKa = 5.37DD91 pKa = 4.32NSLPYY96 pKa = 10.41YY97 pKa = 10.66FEE99 pKa = 4.49GFSYY103 pKa = 10.59SHH105 pKa = 7.06IKK107 pKa = 10.34RR108 pKa = 11.84FLVEE112 pKa = 4.33LADD115 pKa = 3.82KK116 pKa = 9.98FQNRR120 pKa = 11.84YY121 pKa = 7.61HH122 pKa = 6.84TGTSCGVRR130 pKa = 11.84YY131 pKa = 8.46MVASEE136 pKa = 3.77YY137 pKa = 11.07GYY139 pKa = 11.27AEE141 pKa = 4.78EE142 pKa = 4.25YY143 pKa = 10.98LDD145 pKa = 5.34DD146 pKa = 5.04FGHH149 pKa = 6.37SRR151 pKa = 11.84IGQAAPHH158 pKa = 4.95YY159 pKa = 9.69HH160 pKa = 5.85VLLFFPSVAYY170 pKa = 10.23NDD172 pKa = 3.36KK173 pKa = 10.97DD174 pKa = 3.38YY175 pKa = 10.74WADD178 pKa = 3.39VVEE181 pKa = 4.65SLWSVTHH188 pKa = 4.55RR189 pKa = 11.84HH190 pKa = 5.57GRR192 pKa = 11.84VGWSKK197 pKa = 10.69KK198 pKa = 9.07HH199 pKa = 5.4GKK201 pKa = 9.44FVTSPLAIKK210 pKa = 9.16YY211 pKa = 5.33TTQYY215 pKa = 10.16CHH217 pKa = 6.53KK218 pKa = 9.46QHH220 pKa = 6.74SYY222 pKa = 7.52YY223 pKa = 10.24QRR225 pKa = 11.84PEE227 pKa = 3.64IEE229 pKa = 3.49KK230 pKa = 9.78WLYY233 pKa = 10.58QDD235 pKa = 3.29ITKK238 pKa = 8.14DD239 pKa = 3.52TPFHH243 pKa = 5.72VQHH246 pKa = 6.29EE247 pKa = 4.63LIKK250 pKa = 10.47EE251 pKa = 4.07RR252 pKa = 11.84EE253 pKa = 4.11KK254 pKa = 10.86QIHH257 pKa = 7.4DD258 pKa = 3.31ILPFHH263 pKa = 7.23RR264 pKa = 11.84EE265 pKa = 3.69SLHH268 pKa = 7.05FGEE271 pKa = 5.38CAVDD275 pKa = 4.0FLKK278 pKa = 11.04SLSSEE283 pKa = 3.9EE284 pKa = 4.15RR285 pKa = 11.84IKK287 pKa = 11.09LLSNGITFPDD297 pKa = 4.07DD298 pKa = 3.3FAADD302 pKa = 3.67GTLNHH307 pKa = 6.88YY308 pKa = 10.15ILPKK312 pKa = 10.23YY313 pKa = 10.33IYY315 pKa = 9.78DD316 pKa = 3.4KK317 pKa = 11.09LYY319 pKa = 10.58FVIEE323 pKa = 4.16NKK325 pKa = 10.42SVIEE329 pKa = 4.35DD330 pKa = 3.43GKK332 pKa = 11.02SVVKK336 pKa = 10.07PFVRR340 pKa = 11.84YY341 pKa = 9.92GSDD344 pKa = 3.27YY345 pKa = 11.65ALADD349 pKa = 3.35YY350 pKa = 7.5RR351 pKa = 11.84TLFRR355 pKa = 11.84YY356 pKa = 9.22NFNKK360 pKa = 10.11SRR362 pKa = 11.84FDD364 pKa = 3.72LEE366 pKa = 5.28RR367 pKa = 11.84ITSNLFLSQLDD378 pKa = 3.56KK379 pKa = 11.47EE380 pKa = 4.95LIEE383 pKa = 4.45KK384 pKa = 10.13LNKK387 pKa = 9.16RR388 pKa = 11.84DD389 pKa = 3.66YY390 pKa = 10.6VLQYY394 pKa = 10.17NYY396 pKa = 11.17ADD398 pKa = 3.56FRR400 pKa = 11.84HH401 pKa = 6.53FIDD404 pKa = 5.65DD405 pKa = 3.14IKK407 pKa = 11.32SRR409 pKa = 11.84YY410 pKa = 7.3TFDD413 pKa = 5.63RR414 pKa = 11.84ILSYY418 pKa = 10.96KK419 pKa = 10.3HH420 pKa = 6.13IVHH423 pKa = 6.71HH424 pKa = 6.63LFAFDD429 pKa = 3.95PLSYY433 pKa = 11.1SDD435 pKa = 4.92DD436 pKa = 3.57DD437 pKa = 4.52LYY439 pKa = 11.96NFVEE443 pKa = 5.13DD444 pKa = 3.44ISVNKK449 pKa = 10.13ILTKK453 pKa = 10.43RR454 pKa = 11.84FSSLDD459 pKa = 3.33KK460 pKa = 11.23YY461 pKa = 10.82EE462 pKa = 3.92LSIYY466 pKa = 10.29QKK468 pKa = 10.88LFSSYY473 pKa = 10.17NSPDD477 pKa = 2.91ISYY480 pKa = 10.9LDD482 pKa = 4.28GYY484 pKa = 10.75SCVRR488 pKa = 11.84NDD490 pKa = 3.89EE491 pKa = 4.17LDD493 pKa = 3.54EE494 pKa = 4.34FLSIIDD500 pKa = 4.24TIKK503 pKa = 10.62LHH505 pKa = 7.11ASVHH509 pKa = 5.53RR510 pKa = 11.84KK511 pKa = 8.77KK512 pKa = 10.8AYY514 pKa = 10.36DD515 pKa = 3.66LVCLQKK521 pKa = 10.3QAHH524 pKa = 7.17KK525 pKa = 10.57YY526 pKa = 9.84RR527 pKa = 11.84NLSKK531 pKa = 11.2YY532 pKa = 10.22ILL534 pKa = 3.91
Molecular weight: 63.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1658 |
107 |
668 |
414.5 |
47.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.514 ± 1.424 | 1.086 ± 0.392 |
7.057 ± 0.594 | 4.524 ± 0.944 |
5.127 ± 1.063 | 5.127 ± 0.766 |
2.473 ± 0.84 | 6.152 ± 0.562 |
5.489 ± 0.931 | 9.409 ± 0.742 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.87 ± 0.438 | 5.368 ± 0.439 |
3.739 ± 0.882 | 4.222 ± 0.559 |
4.584 ± 0.469 | 9.59 ± 0.631 |
5.79 ± 0.891 | 5.006 ± 0.245 |
1.086 ± 0.231 | 5.79 ± 1.067 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
