
Torque teno virus 16
Taxonomy: Viruses; Anelloviridae; Alphatorquevirus
Average proteome isoelectric point is 7.91
Get precalculated fractions of proteins
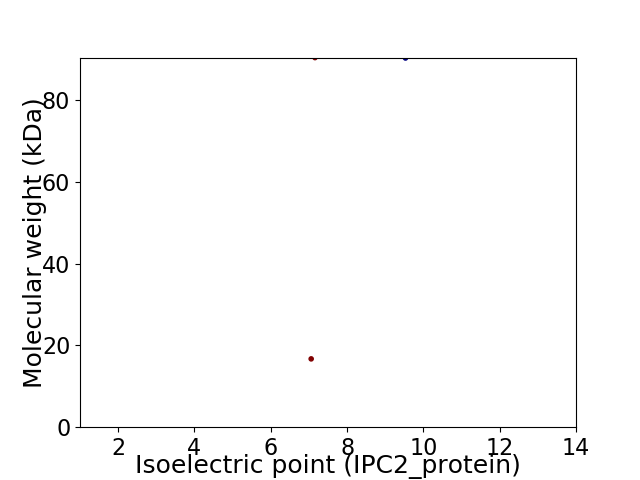
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9WT89|Q9WT89_9VIRU Uncharacterized protein OS=Torque teno virus 16 OX=687355 PE=4 SV=1
MM1 pKa = 7.31HH2 pKa = 7.27FRR4 pKa = 11.84RR5 pKa = 11.84VRR7 pKa = 11.84AKK9 pKa = 10.52RR10 pKa = 11.84KK11 pKa = 9.61LLLQAVRR18 pKa = 11.84APPKK22 pKa = 10.08APAMSFTTPTINAGIRR38 pKa = 11.84EE39 pKa = 4.24QQWFEE44 pKa = 3.86STLRR48 pKa = 11.84SHH50 pKa = 7.42HH51 pKa = 6.01SFCGCGDD58 pKa = 3.78PVLHH62 pKa = 6.0FTNLATRR69 pKa = 11.84FNYY72 pKa = 10.2LPATSSPLDD81 pKa = 3.66PPGPAPRR88 pKa = 11.84GRR90 pKa = 11.84PALRR94 pKa = 11.84RR95 pKa = 11.84LPALPSAPATPSRR108 pKa = 11.84EE109 pKa = 4.27LAWPTGSEE117 pKa = 4.04GGAGGRR123 pKa = 11.84GAGGEE128 pKa = 4.0GGAAVEE134 pKa = 3.77GDD136 pKa = 3.71YY137 pKa = 11.18RR138 pKa = 11.84EE139 pKa = 4.42EE140 pKa = 3.95EE141 pKa = 4.16LDD143 pKa = 3.61EE144 pKa = 5.13LFAALEE150 pKa = 3.98EE151 pKa = 4.76DD152 pKa = 3.98ANQGG156 pKa = 3.26
MM1 pKa = 7.31HH2 pKa = 7.27FRR4 pKa = 11.84RR5 pKa = 11.84VRR7 pKa = 11.84AKK9 pKa = 10.52RR10 pKa = 11.84KK11 pKa = 9.61LLLQAVRR18 pKa = 11.84APPKK22 pKa = 10.08APAMSFTTPTINAGIRR38 pKa = 11.84EE39 pKa = 4.24QQWFEE44 pKa = 3.86STLRR48 pKa = 11.84SHH50 pKa = 7.42HH51 pKa = 6.01SFCGCGDD58 pKa = 3.78PVLHH62 pKa = 6.0FTNLATRR69 pKa = 11.84FNYY72 pKa = 10.2LPATSSPLDD81 pKa = 3.66PPGPAPRR88 pKa = 11.84GRR90 pKa = 11.84PALRR94 pKa = 11.84RR95 pKa = 11.84LPALPSAPATPSRR108 pKa = 11.84EE109 pKa = 4.27LAWPTGSEE117 pKa = 4.04GGAGGRR123 pKa = 11.84GAGGEE128 pKa = 4.0GGAAVEE134 pKa = 3.77GDD136 pKa = 3.71YY137 pKa = 11.18RR138 pKa = 11.84EE139 pKa = 4.42EE140 pKa = 3.95EE141 pKa = 4.16LDD143 pKa = 3.61EE144 pKa = 5.13LFAALEE150 pKa = 3.98EE151 pKa = 4.76DD152 pKa = 3.98ANQGG156 pKa = 3.26
Molecular weight: 16.71 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9WT89|Q9WT89_9VIRU Uncharacterized protein OS=Torque teno virus 16 OX=687355 PE=4 SV=1
MM1 pKa = 7.69AYY3 pKa = 9.45WFRR6 pKa = 11.84RR7 pKa = 11.84WGWRR11 pKa = 11.84PRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84WRR17 pKa = 11.84RR18 pKa = 11.84WRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84LPRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84TRR32 pKa = 11.84RR33 pKa = 11.84AVRR36 pKa = 11.84GLGRR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84KK43 pKa = 8.52PRR45 pKa = 11.84VRR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84TRR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84TYY57 pKa = 9.09RR58 pKa = 11.84RR59 pKa = 11.84GWRR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84YY66 pKa = 8.88IRR68 pKa = 11.84RR69 pKa = 11.84GRR71 pKa = 11.84RR72 pKa = 11.84KK73 pKa = 9.57KK74 pKa = 10.68KK75 pKa = 10.72LILTQWNPAIVKK87 pKa = 9.7RR88 pKa = 11.84CNIKK92 pKa = 10.51GGLPIIICGEE102 pKa = 3.45PRR104 pKa = 11.84AAFNYY109 pKa = 8.68GYY111 pKa = 10.75HH112 pKa = 5.92MEE114 pKa = 5.81DD115 pKa = 3.7YY116 pKa = 9.9TPQPFPFGGGMSTVTFSLKK135 pKa = 10.53ALYY138 pKa = 9.74DD139 pKa = 3.48QYY141 pKa = 11.65LKK143 pKa = 10.71HH144 pKa = 5.79QNRR147 pKa = 11.84WTFSNDD153 pKa = 3.06QLDD156 pKa = 3.68LARR159 pKa = 11.84YY160 pKa = 8.68RR161 pKa = 11.84GCKK164 pKa = 9.25LRR166 pKa = 11.84FYY168 pKa = 10.51RR169 pKa = 11.84SPVCDD174 pKa = 5.25FIVHH178 pKa = 5.98YY179 pKa = 10.89NLIPPLKK186 pKa = 8.69MNQFTSPNTHH196 pKa = 7.18PGLLMLSKK204 pKa = 10.44HH205 pKa = 6.3KK206 pKa = 10.49IIIPSFQTRR215 pKa = 11.84PGGRR219 pKa = 11.84RR220 pKa = 11.84FVKK223 pKa = 10.22IRR225 pKa = 11.84LNPPKK230 pKa = 10.41LFEE233 pKa = 5.13DD234 pKa = 2.98KK235 pKa = 10.34WYY237 pKa = 8.62TQQDD241 pKa = 3.83LCKK244 pKa = 9.78VPLVSITATAADD256 pKa = 4.19LRR258 pKa = 11.84YY259 pKa = 9.18PFCSPQTNNPCTTFQVLRR277 pKa = 11.84KK278 pKa = 9.91NYY280 pKa = 8.07NTVIGTSVKK289 pKa = 10.56DD290 pKa = 3.54QEE292 pKa = 4.49STQDD296 pKa = 3.5FEE298 pKa = 3.92NWLYY302 pKa = 9.76KK303 pKa = 9.89TDD305 pKa = 3.03SHH307 pKa = 6.42YY308 pKa = 10.39QTFATEE314 pKa = 3.96AQLGRR319 pKa = 11.84IPAFNPDD326 pKa = 2.79GTKK329 pKa = 8.63NTKK332 pKa = 7.63QQSWQDD338 pKa = 2.97NWSKK342 pKa = 11.3KK343 pKa = 8.84NSPWTGNSGTYY354 pKa = 9.49PQTTSEE360 pKa = 4.38MYY362 pKa = 10.39KK363 pKa = 9.91IPYY366 pKa = 9.37DD367 pKa = 3.94SNFGFPTYY375 pKa = 10.1RR376 pKa = 11.84AQKK379 pKa = 10.54DD380 pKa = 3.76YY381 pKa = 10.94ILEE384 pKa = 4.02RR385 pKa = 11.84RR386 pKa = 11.84QCNFNYY392 pKa = 9.97EE393 pKa = 4.19VNNPVSKK400 pKa = 10.61KK401 pKa = 8.8VWPQPSTTTPTVDD414 pKa = 3.1YY415 pKa = 11.28YY416 pKa = 10.88EE417 pKa = 4.32YY418 pKa = 10.56HH419 pKa = 6.83CGWFSNIFIGPNRR432 pKa = 11.84YY433 pKa = 8.84NLQFQTAYY441 pKa = 11.16VDD443 pKa = 3.42TTYY446 pKa = 11.39NPLMDD451 pKa = 4.38KK452 pKa = 11.3GKK454 pKa = 10.94GNKK457 pKa = 8.5IWFQYY462 pKa = 10.44LSKK465 pKa = 10.86KK466 pKa = 8.44GTDD469 pKa = 3.68YY470 pKa = 11.58NEE472 pKa = 4.07KK473 pKa = 10.0QCYY476 pKa = 7.4CTLEE480 pKa = 4.98DD481 pKa = 3.49MPLWAICFGYY491 pKa = 9.13TDD493 pKa = 3.69YY494 pKa = 11.82VEE496 pKa = 4.43TQLGPNVDD504 pKa = 3.97HH505 pKa = 6.54EE506 pKa = 4.62TAGLIIMICPYY517 pKa = 8.32TQPPMYY523 pKa = 10.47DD524 pKa = 2.92KK525 pKa = 11.11NRR527 pKa = 11.84PNWGYY532 pKa = 10.95VVYY535 pKa = 8.77DD536 pKa = 3.67TNFGNGKK543 pKa = 8.25MPSGSGQVPVYY554 pKa = 8.06WQCRR558 pKa = 11.84WRR560 pKa = 11.84PMLWFQQQVLNDD572 pKa = 3.43ISKK575 pKa = 8.41TGPYY579 pKa = 10.05AYY581 pKa = 9.58RR582 pKa = 11.84DD583 pKa = 3.63EE584 pKa = 4.51YY585 pKa = 11.78KK586 pKa = 10.64NVQLTLYY593 pKa = 10.85YY594 pKa = 11.01NFIFNWGGDD603 pKa = 3.3MYY605 pKa = 11.24YY606 pKa = 10.04PQVVKK611 pKa = 10.78NPCGDD616 pKa = 3.39SGIVPGSGRR625 pKa = 11.84FTRR628 pKa = 11.84EE629 pKa = 3.72VQVVSPLSMGPAYY642 pKa = 9.7IFHH645 pKa = 6.65YY646 pKa = 10.07FDD648 pKa = 3.05SRR650 pKa = 11.84RR651 pKa = 11.84GFFSEE656 pKa = 4.26KK657 pKa = 9.91ALKK660 pKa = 10.44RR661 pKa = 11.84MQQQQEE667 pKa = 3.86FDD669 pKa = 3.17EE670 pKa = 4.97SFTFKK675 pKa = 10.57PKK677 pKa = 10.19RR678 pKa = 11.84PKK680 pKa = 10.68LSTAAAEE687 pKa = 4.0ILQLEE692 pKa = 4.4EE693 pKa = 5.31DD694 pKa = 3.97STSGEE699 pKa = 4.22GKK701 pKa = 10.26SPLQQEE707 pKa = 4.37EE708 pKa = 4.78KK709 pKa = 10.31EE710 pKa = 4.42VEE712 pKa = 4.28VLQTPTVQLQLQRR725 pKa = 11.84NIQEE729 pKa = 3.88QLAIKK734 pKa = 9.97QQLQFLLLQLLKK746 pKa = 9.5TQSNLHH752 pKa = 6.52LNPQFLSPSS761 pKa = 3.4
MM1 pKa = 7.69AYY3 pKa = 9.45WFRR6 pKa = 11.84RR7 pKa = 11.84WGWRR11 pKa = 11.84PRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84WRR17 pKa = 11.84RR18 pKa = 11.84WRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84LPRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84TRR32 pKa = 11.84RR33 pKa = 11.84AVRR36 pKa = 11.84GLGRR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84KK43 pKa = 8.52PRR45 pKa = 11.84VRR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84TRR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84TYY57 pKa = 9.09RR58 pKa = 11.84RR59 pKa = 11.84GWRR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84YY66 pKa = 8.88IRR68 pKa = 11.84RR69 pKa = 11.84GRR71 pKa = 11.84RR72 pKa = 11.84KK73 pKa = 9.57KK74 pKa = 10.68KK75 pKa = 10.72LILTQWNPAIVKK87 pKa = 9.7RR88 pKa = 11.84CNIKK92 pKa = 10.51GGLPIIICGEE102 pKa = 3.45PRR104 pKa = 11.84AAFNYY109 pKa = 8.68GYY111 pKa = 10.75HH112 pKa = 5.92MEE114 pKa = 5.81DD115 pKa = 3.7YY116 pKa = 9.9TPQPFPFGGGMSTVTFSLKK135 pKa = 10.53ALYY138 pKa = 9.74DD139 pKa = 3.48QYY141 pKa = 11.65LKK143 pKa = 10.71HH144 pKa = 5.79QNRR147 pKa = 11.84WTFSNDD153 pKa = 3.06QLDD156 pKa = 3.68LARR159 pKa = 11.84YY160 pKa = 8.68RR161 pKa = 11.84GCKK164 pKa = 9.25LRR166 pKa = 11.84FYY168 pKa = 10.51RR169 pKa = 11.84SPVCDD174 pKa = 5.25FIVHH178 pKa = 5.98YY179 pKa = 10.89NLIPPLKK186 pKa = 8.69MNQFTSPNTHH196 pKa = 7.18PGLLMLSKK204 pKa = 10.44HH205 pKa = 6.3KK206 pKa = 10.49IIIPSFQTRR215 pKa = 11.84PGGRR219 pKa = 11.84RR220 pKa = 11.84FVKK223 pKa = 10.22IRR225 pKa = 11.84LNPPKK230 pKa = 10.41LFEE233 pKa = 5.13DD234 pKa = 2.98KK235 pKa = 10.34WYY237 pKa = 8.62TQQDD241 pKa = 3.83LCKK244 pKa = 9.78VPLVSITATAADD256 pKa = 4.19LRR258 pKa = 11.84YY259 pKa = 9.18PFCSPQTNNPCTTFQVLRR277 pKa = 11.84KK278 pKa = 9.91NYY280 pKa = 8.07NTVIGTSVKK289 pKa = 10.56DD290 pKa = 3.54QEE292 pKa = 4.49STQDD296 pKa = 3.5FEE298 pKa = 3.92NWLYY302 pKa = 9.76KK303 pKa = 9.89TDD305 pKa = 3.03SHH307 pKa = 6.42YY308 pKa = 10.39QTFATEE314 pKa = 3.96AQLGRR319 pKa = 11.84IPAFNPDD326 pKa = 2.79GTKK329 pKa = 8.63NTKK332 pKa = 7.63QQSWQDD338 pKa = 2.97NWSKK342 pKa = 11.3KK343 pKa = 8.84NSPWTGNSGTYY354 pKa = 9.49PQTTSEE360 pKa = 4.38MYY362 pKa = 10.39KK363 pKa = 9.91IPYY366 pKa = 9.37DD367 pKa = 3.94SNFGFPTYY375 pKa = 10.1RR376 pKa = 11.84AQKK379 pKa = 10.54DD380 pKa = 3.76YY381 pKa = 10.94ILEE384 pKa = 4.02RR385 pKa = 11.84RR386 pKa = 11.84QCNFNYY392 pKa = 9.97EE393 pKa = 4.19VNNPVSKK400 pKa = 10.61KK401 pKa = 8.8VWPQPSTTTPTVDD414 pKa = 3.1YY415 pKa = 11.28YY416 pKa = 10.88EE417 pKa = 4.32YY418 pKa = 10.56HH419 pKa = 6.83CGWFSNIFIGPNRR432 pKa = 11.84YY433 pKa = 8.84NLQFQTAYY441 pKa = 11.16VDD443 pKa = 3.42TTYY446 pKa = 11.39NPLMDD451 pKa = 4.38KK452 pKa = 11.3GKK454 pKa = 10.94GNKK457 pKa = 8.5IWFQYY462 pKa = 10.44LSKK465 pKa = 10.86KK466 pKa = 8.44GTDD469 pKa = 3.68YY470 pKa = 11.58NEE472 pKa = 4.07KK473 pKa = 10.0QCYY476 pKa = 7.4CTLEE480 pKa = 4.98DD481 pKa = 3.49MPLWAICFGYY491 pKa = 9.13TDD493 pKa = 3.69YY494 pKa = 11.82VEE496 pKa = 4.43TQLGPNVDD504 pKa = 3.97HH505 pKa = 6.54EE506 pKa = 4.62TAGLIIMICPYY517 pKa = 8.32TQPPMYY523 pKa = 10.47DD524 pKa = 2.92KK525 pKa = 11.11NRR527 pKa = 11.84PNWGYY532 pKa = 10.95VVYY535 pKa = 8.77DD536 pKa = 3.67TNFGNGKK543 pKa = 8.25MPSGSGQVPVYY554 pKa = 8.06WQCRR558 pKa = 11.84WRR560 pKa = 11.84PMLWFQQQVLNDD572 pKa = 3.43ISKK575 pKa = 8.41TGPYY579 pKa = 10.05AYY581 pKa = 9.58RR582 pKa = 11.84DD583 pKa = 3.63EE584 pKa = 4.51YY585 pKa = 11.78KK586 pKa = 10.64NVQLTLYY593 pKa = 10.85YY594 pKa = 11.01NFIFNWGGDD603 pKa = 3.3MYY605 pKa = 11.24YY606 pKa = 10.04PQVVKK611 pKa = 10.78NPCGDD616 pKa = 3.39SGIVPGSGRR625 pKa = 11.84FTRR628 pKa = 11.84EE629 pKa = 3.72VQVVSPLSMGPAYY642 pKa = 9.7IFHH645 pKa = 6.65YY646 pKa = 10.07FDD648 pKa = 3.05SRR650 pKa = 11.84RR651 pKa = 11.84GFFSEE656 pKa = 4.26KK657 pKa = 9.91ALKK660 pKa = 10.44RR661 pKa = 11.84MQQQQEE667 pKa = 3.86FDD669 pKa = 3.17EE670 pKa = 4.97SFTFKK675 pKa = 10.57PKK677 pKa = 10.19RR678 pKa = 11.84PKK680 pKa = 10.68LSTAAAEE687 pKa = 4.0ILQLEE692 pKa = 4.4EE693 pKa = 5.31DD694 pKa = 3.97STSGEE699 pKa = 4.22GKK701 pKa = 10.26SPLQQEE707 pKa = 4.37EE708 pKa = 4.78KK709 pKa = 10.31EE710 pKa = 4.42VEE712 pKa = 4.28VLQTPTVQLQLQRR725 pKa = 11.84NIQEE729 pKa = 3.88QLAIKK734 pKa = 9.97QQLQFLLLQLLKK746 pKa = 9.5TQSNLHH752 pKa = 6.52LNPQFLSPSS761 pKa = 3.4
Molecular weight: 90.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
917 |
156 |
761 |
458.5 |
53.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.907 ± 4.253 | 1.854 ± 0.284 |
3.926 ± 0.358 | 4.362 ± 1.656 |
5.016 ± 0.263 | 6.87 ± 2.002 |
1.527 ± 0.516 | 3.708 ± 1.206 |
5.453 ± 1.755 | 7.634 ± 0.985 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.854 ± 0.284 | 5.125 ± 1.273 |
7.852 ± 1.514 | 6.434 ± 1.924 |
9.378 ± 0.118 | 5.234 ± 0.266 |
6.761 ± 0.493 | 4.144 ± 0.785 |
2.617 ± 0.664 | 5.344 ± 2.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
