
Bat paramyxovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; unclassified Paramyxoviridae
Average proteome isoelectric point is 6.95
Get precalculated fractions of proteins
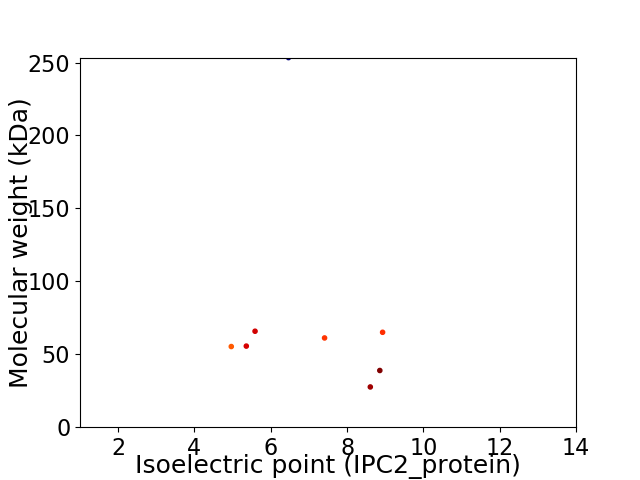
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
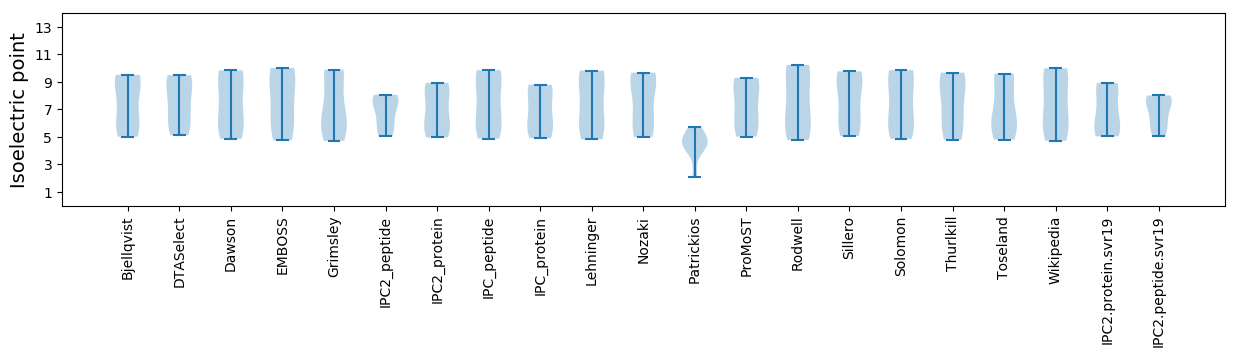
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346NTM2|A0A346NTM2_9MONO Matrix protein OS=Bat paramyxovirus OX=1300978 GN=M PE=4 SV=1
MM1 pKa = 7.34SRR3 pKa = 11.84VDD5 pKa = 3.85KK6 pKa = 11.23LSASVDD12 pKa = 3.19NGLAIAEE19 pKa = 4.73FIQEE23 pKa = 4.14NRR25 pKa = 11.84SSIQSTYY32 pKa = 9.5GRR34 pKa = 11.84SSIGEE39 pKa = 3.66PTIANRR45 pKa = 11.84AKK47 pKa = 10.42AWEE50 pKa = 4.3LYY52 pKa = 9.96IDD54 pKa = 3.71RR55 pKa = 11.84TNSRR59 pKa = 11.84NAGHH63 pKa = 6.45EE64 pKa = 4.39GKK66 pKa = 10.48LKK68 pKa = 10.69SINEE72 pKa = 3.98LPEE75 pKa = 4.44GKK77 pKa = 10.33GSEE80 pKa = 4.22TTRR83 pKa = 11.84VAEE86 pKa = 4.04TSEE89 pKa = 4.4IEE91 pKa = 4.63QSPKK95 pKa = 10.12NQNSSSGDD103 pKa = 3.23GSGDD107 pKa = 3.44FNFRR111 pKa = 11.84FPGDD115 pKa = 3.5SSEE118 pKa = 4.31SDD120 pKa = 2.72IGVWSKK126 pKa = 11.41NSSDD130 pKa = 3.58LSDD133 pKa = 3.47VEE135 pKa = 4.4GRR137 pKa = 11.84SISISEE143 pKa = 4.32GEE145 pKa = 4.42AEE147 pKa = 4.54PEE149 pKa = 4.09TSNDD153 pKa = 2.77VPSQVNEE160 pKa = 3.89SSRR163 pKa = 11.84DD164 pKa = 3.58HH165 pKa = 6.59ARR167 pKa = 11.84EE168 pKa = 3.91GADD171 pKa = 3.26QADD174 pKa = 4.08EE175 pKa = 4.2QVVSSNDD182 pKa = 3.25QEE184 pKa = 4.32TTTIIEE190 pKa = 4.34NPSKK194 pKa = 10.83EE195 pKa = 4.34SIASIMEE202 pKa = 3.93EE203 pKa = 4.09SGPMPKK209 pKa = 9.51RR210 pKa = 11.84RR211 pKa = 11.84LKK213 pKa = 9.35NASRR217 pKa = 11.84IKK219 pKa = 9.49EE220 pKa = 4.32TVDD223 pKa = 4.76AIPADD228 pKa = 3.77TPIVKK233 pKa = 8.68KK234 pKa = 7.56TTGEE238 pKa = 3.99NIQLQKK244 pKa = 10.3KK245 pKa = 6.43TDD247 pKa = 3.67KK248 pKa = 11.42LEE250 pKa = 4.31LSAGATQSAHH260 pKa = 7.28PSPQIQSSTNADD272 pKa = 2.59VDD274 pKa = 3.9TAQDD278 pKa = 3.38HH279 pKa = 4.18VHH281 pKa = 5.35YY282 pKa = 10.88AKK284 pKa = 8.46MTEE287 pKa = 4.12EE288 pKa = 4.04SHH290 pKa = 6.87PLSKK294 pKa = 10.01TDD296 pKa = 3.38TNQIALEE303 pKa = 4.17KK304 pKa = 10.48KK305 pKa = 9.88LDD307 pKa = 4.31KK308 pKa = 10.8ILEE311 pKa = 4.22NQEE314 pKa = 4.45KK315 pKa = 9.91ILKK318 pKa = 9.24KK319 pKa = 10.72LEE321 pKa = 3.9VLSEE325 pKa = 4.1VKK327 pKa = 10.84EE328 pKa = 3.89EE329 pKa = 3.84VNAIKK334 pKa = 9.21KK335 pKa = 7.37TLSNQSLALSTIEE348 pKa = 4.15NYY350 pKa = 9.67IGEE353 pKa = 4.37MMVIIPKK360 pKa = 10.31SGVPEE365 pKa = 4.37DD366 pKa = 4.17EE367 pKa = 4.49NNHH370 pKa = 6.65KK371 pKa = 8.75ITKK374 pKa = 10.19NPDD377 pKa = 2.86LRR379 pKa = 11.84PVIGRR384 pKa = 11.84DD385 pKa = 3.01STRR388 pKa = 11.84GKK390 pKa = 10.57KK391 pKa = 9.22EE392 pKa = 3.83VIRR395 pKa = 11.84PMTDD399 pKa = 2.83RR400 pKa = 11.84EE401 pKa = 4.21RR402 pKa = 11.84IDD404 pKa = 3.32IDD406 pKa = 3.43EE407 pKa = 4.84DD408 pKa = 4.64FYY410 pKa = 11.59NIPVVDD416 pKa = 3.8EE417 pKa = 4.69KK418 pKa = 11.74YY419 pKa = 10.11FVKK422 pKa = 10.6PINKK426 pKa = 9.14QEE428 pKa = 3.66NNAANFVPKK437 pKa = 9.9EE438 pKa = 4.14DD439 pKa = 3.51KK440 pKa = 9.86TSYY443 pKa = 10.54RR444 pKa = 11.84IIRR447 pKa = 11.84DD448 pKa = 3.83IIRR451 pKa = 11.84KK452 pKa = 7.2EE453 pKa = 4.09VKK455 pKa = 10.39DD456 pKa = 3.78PEE458 pKa = 4.49ARR460 pKa = 11.84DD461 pKa = 3.57AVIKK465 pKa = 10.41LFNDD469 pKa = 4.02SIGKK473 pKa = 9.28VPINEE478 pKa = 4.05IYY480 pKa = 10.67DD481 pKa = 4.48GIRR484 pKa = 11.84MLVYY488 pKa = 10.47TDD490 pKa = 4.09ILDD493 pKa = 4.23HH494 pKa = 6.94LEE496 pKa = 4.0KK497 pKa = 10.92
MM1 pKa = 7.34SRR3 pKa = 11.84VDD5 pKa = 3.85KK6 pKa = 11.23LSASVDD12 pKa = 3.19NGLAIAEE19 pKa = 4.73FIQEE23 pKa = 4.14NRR25 pKa = 11.84SSIQSTYY32 pKa = 9.5GRR34 pKa = 11.84SSIGEE39 pKa = 3.66PTIANRR45 pKa = 11.84AKK47 pKa = 10.42AWEE50 pKa = 4.3LYY52 pKa = 9.96IDD54 pKa = 3.71RR55 pKa = 11.84TNSRR59 pKa = 11.84NAGHH63 pKa = 6.45EE64 pKa = 4.39GKK66 pKa = 10.48LKK68 pKa = 10.69SINEE72 pKa = 3.98LPEE75 pKa = 4.44GKK77 pKa = 10.33GSEE80 pKa = 4.22TTRR83 pKa = 11.84VAEE86 pKa = 4.04TSEE89 pKa = 4.4IEE91 pKa = 4.63QSPKK95 pKa = 10.12NQNSSSGDD103 pKa = 3.23GSGDD107 pKa = 3.44FNFRR111 pKa = 11.84FPGDD115 pKa = 3.5SSEE118 pKa = 4.31SDD120 pKa = 2.72IGVWSKK126 pKa = 11.41NSSDD130 pKa = 3.58LSDD133 pKa = 3.47VEE135 pKa = 4.4GRR137 pKa = 11.84SISISEE143 pKa = 4.32GEE145 pKa = 4.42AEE147 pKa = 4.54PEE149 pKa = 4.09TSNDD153 pKa = 2.77VPSQVNEE160 pKa = 3.89SSRR163 pKa = 11.84DD164 pKa = 3.58HH165 pKa = 6.59ARR167 pKa = 11.84EE168 pKa = 3.91GADD171 pKa = 3.26QADD174 pKa = 4.08EE175 pKa = 4.2QVVSSNDD182 pKa = 3.25QEE184 pKa = 4.32TTTIIEE190 pKa = 4.34NPSKK194 pKa = 10.83EE195 pKa = 4.34SIASIMEE202 pKa = 3.93EE203 pKa = 4.09SGPMPKK209 pKa = 9.51RR210 pKa = 11.84RR211 pKa = 11.84LKK213 pKa = 9.35NASRR217 pKa = 11.84IKK219 pKa = 9.49EE220 pKa = 4.32TVDD223 pKa = 4.76AIPADD228 pKa = 3.77TPIVKK233 pKa = 8.68KK234 pKa = 7.56TTGEE238 pKa = 3.99NIQLQKK244 pKa = 10.3KK245 pKa = 6.43TDD247 pKa = 3.67KK248 pKa = 11.42LEE250 pKa = 4.31LSAGATQSAHH260 pKa = 7.28PSPQIQSSTNADD272 pKa = 2.59VDD274 pKa = 3.9TAQDD278 pKa = 3.38HH279 pKa = 4.18VHH281 pKa = 5.35YY282 pKa = 10.88AKK284 pKa = 8.46MTEE287 pKa = 4.12EE288 pKa = 4.04SHH290 pKa = 6.87PLSKK294 pKa = 10.01TDD296 pKa = 3.38TNQIALEE303 pKa = 4.17KK304 pKa = 10.48KK305 pKa = 9.88LDD307 pKa = 4.31KK308 pKa = 10.8ILEE311 pKa = 4.22NQEE314 pKa = 4.45KK315 pKa = 9.91ILKK318 pKa = 9.24KK319 pKa = 10.72LEE321 pKa = 3.9VLSEE325 pKa = 4.1VKK327 pKa = 10.84EE328 pKa = 3.89EE329 pKa = 3.84VNAIKK334 pKa = 9.21KK335 pKa = 7.37TLSNQSLALSTIEE348 pKa = 4.15NYY350 pKa = 9.67IGEE353 pKa = 4.37MMVIIPKK360 pKa = 10.31SGVPEE365 pKa = 4.37DD366 pKa = 4.17EE367 pKa = 4.49NNHH370 pKa = 6.65KK371 pKa = 8.75ITKK374 pKa = 10.19NPDD377 pKa = 2.86LRR379 pKa = 11.84PVIGRR384 pKa = 11.84DD385 pKa = 3.01STRR388 pKa = 11.84GKK390 pKa = 10.57KK391 pKa = 9.22EE392 pKa = 3.83VIRR395 pKa = 11.84PMTDD399 pKa = 2.83RR400 pKa = 11.84EE401 pKa = 4.21RR402 pKa = 11.84IDD404 pKa = 3.32IDD406 pKa = 3.43EE407 pKa = 4.84DD408 pKa = 4.64FYY410 pKa = 11.59NIPVVDD416 pKa = 3.8EE417 pKa = 4.69KK418 pKa = 11.74YY419 pKa = 10.11FVKK422 pKa = 10.6PINKK426 pKa = 9.14QEE428 pKa = 3.66NNAANFVPKK437 pKa = 9.9EE438 pKa = 4.14DD439 pKa = 3.51KK440 pKa = 9.86TSYY443 pKa = 10.54RR444 pKa = 11.84IIRR447 pKa = 11.84DD448 pKa = 3.83IIRR451 pKa = 11.84KK452 pKa = 7.2EE453 pKa = 4.09VKK455 pKa = 10.39DD456 pKa = 3.78PEE458 pKa = 4.49ARR460 pKa = 11.84DD461 pKa = 3.57AVIKK465 pKa = 10.41LFNDD469 pKa = 4.02SIGKK473 pKa = 9.28VPINEE478 pKa = 4.05IYY480 pKa = 10.67DD481 pKa = 4.48GIRR484 pKa = 11.84MLVYY488 pKa = 10.47TDD490 pKa = 4.09ILDD493 pKa = 4.23HH494 pKa = 6.94LEE496 pKa = 4.0KK497 pKa = 10.92
Molecular weight: 55.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346NTM3|A0A346NTM3_9MONO Fusion glycoprotein F0 OS=Bat paramyxovirus OX=1300978 GN=F PE=3 SV=1
MM1 pKa = 7.41SGKK4 pKa = 10.4ADD6 pKa = 3.86FSKK9 pKa = 10.79RR10 pKa = 11.84SWEE13 pKa = 4.59DD14 pKa = 2.85GGTLQAIDD22 pKa = 3.5ATADD26 pKa = 3.3EE27 pKa = 4.94KK28 pKa = 11.46GRR30 pKa = 11.84LIPKK34 pKa = 9.41VRR36 pKa = 11.84VVNPGWNDD44 pKa = 3.34RR45 pKa = 11.84KK46 pKa = 9.51STGYY50 pKa = 10.5KK51 pKa = 9.41YY52 pKa = 10.69LIVYY56 pKa = 7.85GIIEE60 pKa = 4.57EE61 pKa = 4.29KK62 pKa = 10.76EE63 pKa = 3.76KK64 pKa = 11.34DD65 pKa = 3.48HH66 pKa = 5.87THH68 pKa = 5.77SAGGEE73 pKa = 3.93PTVRR77 pKa = 11.84KK78 pKa = 10.1SNAPRR83 pKa = 11.84AFGALPLGVGRR94 pKa = 11.84SHH96 pKa = 7.63VEE98 pKa = 3.78PEE100 pKa = 4.31DD101 pKa = 3.57LLEE104 pKa = 5.44AILKK108 pKa = 10.55LEE110 pKa = 3.69ITVRR114 pKa = 11.84RR115 pKa = 11.84TAGANEE121 pKa = 4.11KK122 pKa = 9.9IVYY125 pKa = 9.43GMSRR129 pKa = 11.84IDD131 pKa = 4.61NILKK135 pKa = 8.97PWAPILEE142 pKa = 4.44NGAIFPAIKK151 pKa = 9.89VCSNIDD157 pKa = 3.72IVPLDD162 pKa = 4.07HH163 pKa = 7.07PLKK166 pKa = 10.41LRR168 pKa = 11.84PIFLTITLLTDD179 pKa = 3.59AGIYY183 pKa = 9.3KK184 pKa = 9.8VPRR187 pKa = 11.84SILEE191 pKa = 3.66FRR193 pKa = 11.84QNKK196 pKa = 9.61AISFNLLVTLQVGSDD211 pKa = 3.73FTSSGVKK218 pKa = 10.56GMINEE223 pKa = 3.93NGEE226 pKa = 4.17RR227 pKa = 11.84VTTFMVHH234 pKa = 5.95IGNFVRR240 pKa = 11.84KK241 pKa = 9.36RR242 pKa = 11.84GKK244 pKa = 9.99AYY246 pKa = 10.54SIEE249 pKa = 4.12YY250 pKa = 10.29CKK252 pKa = 11.04AKK254 pKa = 9.77VDD256 pKa = 3.62KK257 pKa = 10.41MDD259 pKa = 3.27MRR261 pKa = 11.84FSLGAVGGLSFHH273 pKa = 6.22VRR275 pKa = 11.84IAGKK279 pKa = 9.07MSHH282 pKa = 5.92MLKK285 pKa = 10.42AQLGYY290 pKa = 10.63HH291 pKa = 5.19RR292 pKa = 11.84TICYY296 pKa = 10.83SLMDD300 pKa = 4.0TNPYY304 pKa = 9.59LNRR307 pKa = 11.84VMWRR311 pKa = 11.84SEE313 pKa = 3.75CSILKK318 pKa = 8.08VTAVFQPSVPKK329 pKa = 10.26EE330 pKa = 3.51FKK332 pKa = 10.41IYY334 pKa = 10.8DD335 pKa = 3.81DD336 pKa = 3.91VLIDD340 pKa = 3.61NTGKK344 pKa = 10.12ILKK347 pKa = 9.31QQ348 pKa = 3.23
MM1 pKa = 7.41SGKK4 pKa = 10.4ADD6 pKa = 3.86FSKK9 pKa = 10.79RR10 pKa = 11.84SWEE13 pKa = 4.59DD14 pKa = 2.85GGTLQAIDD22 pKa = 3.5ATADD26 pKa = 3.3EE27 pKa = 4.94KK28 pKa = 11.46GRR30 pKa = 11.84LIPKK34 pKa = 9.41VRR36 pKa = 11.84VVNPGWNDD44 pKa = 3.34RR45 pKa = 11.84KK46 pKa = 9.51STGYY50 pKa = 10.5KK51 pKa = 9.41YY52 pKa = 10.69LIVYY56 pKa = 7.85GIIEE60 pKa = 4.57EE61 pKa = 4.29KK62 pKa = 10.76EE63 pKa = 3.76KK64 pKa = 11.34DD65 pKa = 3.48HH66 pKa = 5.87THH68 pKa = 5.77SAGGEE73 pKa = 3.93PTVRR77 pKa = 11.84KK78 pKa = 10.1SNAPRR83 pKa = 11.84AFGALPLGVGRR94 pKa = 11.84SHH96 pKa = 7.63VEE98 pKa = 3.78PEE100 pKa = 4.31DD101 pKa = 3.57LLEE104 pKa = 5.44AILKK108 pKa = 10.55LEE110 pKa = 3.69ITVRR114 pKa = 11.84RR115 pKa = 11.84TAGANEE121 pKa = 4.11KK122 pKa = 9.9IVYY125 pKa = 9.43GMSRR129 pKa = 11.84IDD131 pKa = 4.61NILKK135 pKa = 8.97PWAPILEE142 pKa = 4.44NGAIFPAIKK151 pKa = 9.89VCSNIDD157 pKa = 3.72IVPLDD162 pKa = 4.07HH163 pKa = 7.07PLKK166 pKa = 10.41LRR168 pKa = 11.84PIFLTITLLTDD179 pKa = 3.59AGIYY183 pKa = 9.3KK184 pKa = 9.8VPRR187 pKa = 11.84SILEE191 pKa = 3.66FRR193 pKa = 11.84QNKK196 pKa = 9.61AISFNLLVTLQVGSDD211 pKa = 3.73FTSSGVKK218 pKa = 10.56GMINEE223 pKa = 3.93NGEE226 pKa = 4.17RR227 pKa = 11.84VTTFMVHH234 pKa = 5.95IGNFVRR240 pKa = 11.84KK241 pKa = 9.36RR242 pKa = 11.84GKK244 pKa = 9.99AYY246 pKa = 10.54SIEE249 pKa = 4.12YY250 pKa = 10.29CKK252 pKa = 11.04AKK254 pKa = 9.77VDD256 pKa = 3.62KK257 pKa = 10.41MDD259 pKa = 3.27MRR261 pKa = 11.84FSLGAVGGLSFHH273 pKa = 6.22VRR275 pKa = 11.84IAGKK279 pKa = 9.07MSHH282 pKa = 5.92MLKK285 pKa = 10.42AQLGYY290 pKa = 10.63HH291 pKa = 5.19RR292 pKa = 11.84TICYY296 pKa = 10.83SLMDD300 pKa = 4.0TNPYY304 pKa = 9.59LNRR307 pKa = 11.84VMWRR311 pKa = 11.84SEE313 pKa = 3.75CSILKK318 pKa = 8.08VTAVFQPSVPKK329 pKa = 10.26EE330 pKa = 3.51FKK332 pKa = 10.41IYY334 pKa = 10.8DD335 pKa = 3.81DD336 pKa = 3.91VLIDD340 pKa = 3.61NTGKK344 pKa = 10.12ILKK347 pKa = 9.31QQ348 pKa = 3.23
Molecular weight: 38.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5477 |
239 |
2195 |
684.6 |
77.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.076 ± 0.698 | 1.296 ± 0.288 |
6.08 ± 0.235 | 5.77 ± 0.592 |
3.834 ± 0.446 | 5.094 ± 0.666 |
1.917 ± 0.354 | 8.527 ± 0.438 |
6.865 ± 0.645 | 9.385 ± 0.778 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.666 ± 0.333 | 6.463 ± 0.455 |
3.944 ± 0.233 | 3.688 ± 0.306 |
5.185 ± 0.321 | 8.234 ± 0.57 |
5.879 ± 0.45 | 5.222 ± 0.353 |
1.15 ± 0.121 | 3.725 ± 0.431 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
