
Rhizophagus irregularis (strain DAOM 181602 / DAOM 197198 / MUCL 43194) (Arbuscular mycorrhizal fungus) (Glomus intraradices)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Fungi incertae sedis; Mucoromycota; Glomeromycotina; Glomeromycetes; Glomerales; Glomeraceae; Rhizophagus; Rhizophagus irregularis
Average proteome isoelectric point is 6.85
Get precalculated fractions of proteins
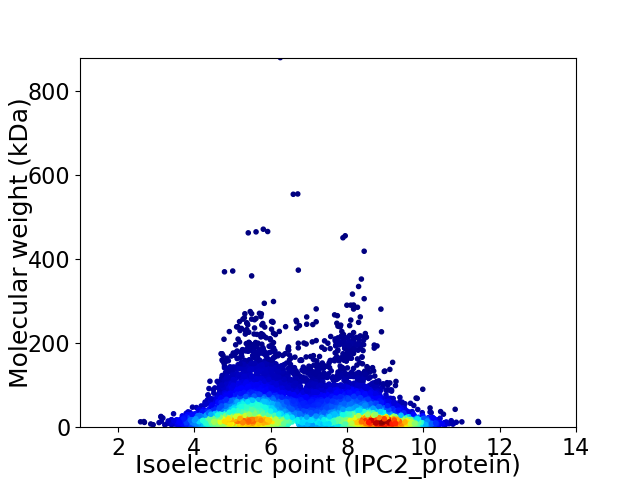
Virtual 2D-PAGE plot for 11211 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H5TAK6|A0A2H5TAK6_RHIID JEMT01018572.1_cds_EXX66627.1_13402: PROVISIONAL OS=Rhizophagus irregularis (strain DAOM 181602 / DAOM 197198 / MUCL 43194) OX=747089 GN=RIR_2333520 PE=4 SV=1
MM1 pKa = 7.67SSNQVEE7 pKa = 4.76MNNLMEE13 pKa = 4.36DD14 pKa = 3.42HH15 pKa = 6.78YY16 pKa = 10.54LNSVIPSVASQPIDD30 pKa = 3.36EE31 pKa = 4.89EE32 pKa = 4.52KK33 pKa = 11.22NEE35 pKa = 3.99NEE37 pKa = 4.33DD38 pKa = 3.67TFSLLNLSNVEE49 pKa = 4.42FEE51 pKa = 4.12YY52 pKa = 10.25TYY54 pKa = 11.59DD55 pKa = 3.53NVIDD59 pKa = 3.95VSLMSMNQTRR69 pKa = 11.84MNTLTEE75 pKa = 4.13EE76 pKa = 4.32VHH78 pKa = 6.7CLNNMIPSVASQPIDD93 pKa = 3.27EE94 pKa = 4.58SLEE97 pKa = 3.95YY98 pKa = 10.17TYY100 pKa = 11.33DD101 pKa = 3.37NQFIIEE107 pKa = 4.32NVSLMSMNQVEE118 pKa = 4.9MNNLMEE124 pKa = 4.36DD125 pKa = 3.42HH126 pKa = 6.78YY127 pKa = 10.54LNSVIPSFASQPIDD141 pKa = 3.51EE142 pKa = 4.87KK143 pKa = 11.77NEE145 pKa = 3.87FPLLNLGLLEE155 pKa = 4.11STYY158 pKa = 11.41DD159 pKa = 3.38NQFIIEE165 pKa = 4.91DD166 pKa = 3.96VPLMPMNQATMNNPIEE182 pKa = 4.37EE183 pKa = 4.2DD184 pKa = 3.16HH185 pKa = 6.7FLNGVIPSFVNQPIYY200 pKa = 10.25EE201 pKa = 4.16RR202 pKa = 11.84KK203 pKa = 10.09NKK205 pKa = 9.81IEE207 pKa = 4.57EE208 pKa = 3.99ISPLHH213 pKa = 5.72LTYY216 pKa = 10.97DD217 pKa = 3.48NQFIIEE223 pKa = 4.59DD224 pKa = 3.68VSLMSVASQPIYY236 pKa = 10.22EE237 pKa = 4.68EE238 pKa = 4.2KK239 pKa = 11.09NKK241 pKa = 11.2DD242 pKa = 3.64EE243 pKa = 4.93VNDD246 pKa = 3.8TDD248 pKa = 5.31DD249 pKa = 4.32VIGSKK254 pKa = 9.63MFDD257 pKa = 3.48FVNN260 pKa = 3.46
MM1 pKa = 7.67SSNQVEE7 pKa = 4.76MNNLMEE13 pKa = 4.36DD14 pKa = 3.42HH15 pKa = 6.78YY16 pKa = 10.54LNSVIPSVASQPIDD30 pKa = 3.36EE31 pKa = 4.89EE32 pKa = 4.52KK33 pKa = 11.22NEE35 pKa = 3.99NEE37 pKa = 4.33DD38 pKa = 3.67TFSLLNLSNVEE49 pKa = 4.42FEE51 pKa = 4.12YY52 pKa = 10.25TYY54 pKa = 11.59DD55 pKa = 3.53NVIDD59 pKa = 3.95VSLMSMNQTRR69 pKa = 11.84MNTLTEE75 pKa = 4.13EE76 pKa = 4.32VHH78 pKa = 6.7CLNNMIPSVASQPIDD93 pKa = 3.27EE94 pKa = 4.58SLEE97 pKa = 3.95YY98 pKa = 10.17TYY100 pKa = 11.33DD101 pKa = 3.37NQFIIEE107 pKa = 4.32NVSLMSMNQVEE118 pKa = 4.9MNNLMEE124 pKa = 4.36DD125 pKa = 3.42HH126 pKa = 6.78YY127 pKa = 10.54LNSVIPSFASQPIDD141 pKa = 3.51EE142 pKa = 4.87KK143 pKa = 11.77NEE145 pKa = 3.87FPLLNLGLLEE155 pKa = 4.11STYY158 pKa = 11.41DD159 pKa = 3.38NQFIIEE165 pKa = 4.91DD166 pKa = 3.96VPLMPMNQATMNNPIEE182 pKa = 4.37EE183 pKa = 4.2DD184 pKa = 3.16HH185 pKa = 6.7FLNGVIPSFVNQPIYY200 pKa = 10.25EE201 pKa = 4.16RR202 pKa = 11.84KK203 pKa = 10.09NKK205 pKa = 9.81IEE207 pKa = 4.57EE208 pKa = 3.99ISPLHH213 pKa = 5.72LTYY216 pKa = 10.97DD217 pKa = 3.48NQFIIEE223 pKa = 4.59DD224 pKa = 3.68VSLMSVASQPIYY236 pKa = 10.22EE237 pKa = 4.68EE238 pKa = 4.2KK239 pKa = 11.09NKK241 pKa = 11.2DD242 pKa = 3.64EE243 pKa = 4.93VNDD246 pKa = 3.8TDD248 pKa = 5.31DD249 pKa = 4.32VIGSKK254 pKa = 9.63MFDD257 pKa = 3.48FVNN260 pKa = 3.46
Molecular weight: 29.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U9SIX8|U9SIX8_RHIID Tis13_13353: PROVISIONAL OS=Rhizophagus irregularis (strain DAOM 181602 / DAOM 197198 / MUCL 43194) OX=747089 GN=GLOINDRAFT_13241 PE=4 SV=1
MM1 pKa = 5.84VTSWLRR7 pKa = 11.84LGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGFFFFLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGFRR107 pKa = 11.84VRR109 pKa = 11.84VRR111 pKa = 11.84VRR113 pKa = 11.84VRR115 pKa = 11.84VRR117 pKa = 11.84VRR119 pKa = 11.84VRR121 pKa = 11.84VRR123 pKa = 11.84VRR125 pKa = 11.84VRR127 pKa = 11.84VRR129 pKa = 11.84VRR131 pKa = 11.84VRR133 pKa = 11.84VRR135 pKa = 11.84VRR137 pKa = 11.84GG138 pKa = 3.53
MM1 pKa = 5.84VTSWLRR7 pKa = 11.84LGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGFFFFLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGFRR107 pKa = 11.84VRR109 pKa = 11.84VRR111 pKa = 11.84VRR113 pKa = 11.84VRR115 pKa = 11.84VRR117 pKa = 11.84VRR119 pKa = 11.84VRR121 pKa = 11.84VRR123 pKa = 11.84VRR125 pKa = 11.84VRR127 pKa = 11.84VRR129 pKa = 11.84VRR131 pKa = 11.84VRR133 pKa = 11.84VRR135 pKa = 11.84VRR137 pKa = 11.84GG138 pKa = 3.53
Molecular weight: 13.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
3814083 |
20 |
7762 |
340.2 |
39.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.518 ± 0.025 | 1.636 ± 0.013 |
5.932 ± 0.017 | 6.868 ± 0.035 |
4.511 ± 0.017 | 4.183 ± 0.021 |
2.306 ± 0.012 | 8.079 ± 0.026 |
7.817 ± 0.034 | 9.064 ± 0.034 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.972 ± 0.01 | 7.15 ± 0.035 |
4.103 ± 0.023 | 3.871 ± 0.019 |
4.738 ± 0.024 | 7.912 ± 0.043 |
5.273 ± 0.019 | 4.669 ± 0.018 |
1.354 ± 0.01 | 4.044 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
