
Thiobacillus denitrificans
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Nitrosomonadales; Thiobacillaceae; Thiobacillus
Average proteome isoelectric point is 6.57
Get precalculated fractions of proteins
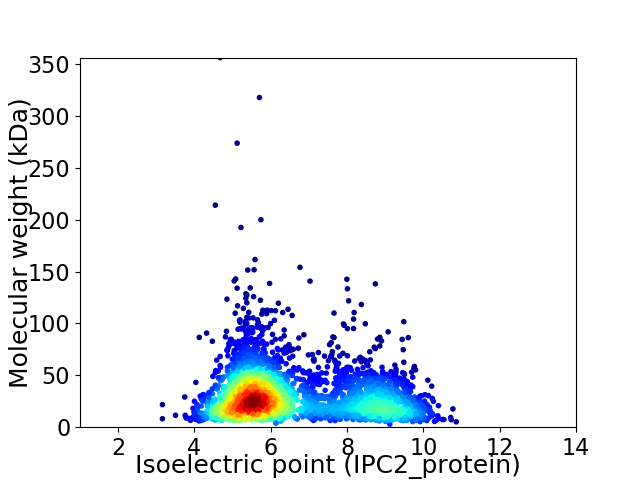
Virtual 2D-PAGE plot for 3078 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
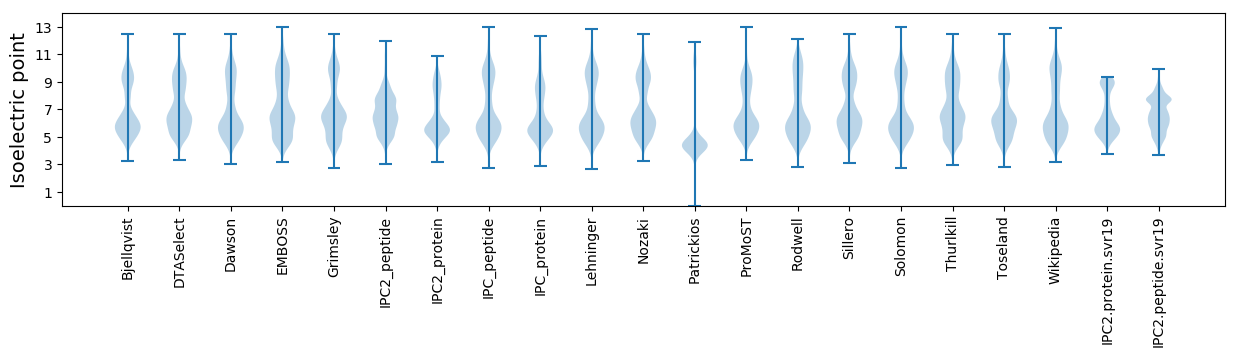
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A106BP43|A0A106BP43_THIDE Protease OS=Thiobacillus denitrificans OX=36861 GN=ABW22_08315 PE=4 SV=1
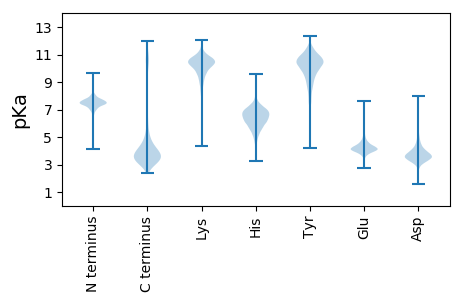
MM1 pKa = 7.33KK2 pKa = 10.1HH3 pKa = 6.93LIPEE7 pKa = 4.57TPPDD11 pKa = 3.76YY12 pKa = 11.24DD13 pKa = 3.14HH14 pKa = 6.79TRR16 pKa = 11.84IIEE19 pKa = 4.29RR20 pKa = 11.84PDD22 pKa = 2.75GCYY25 pKa = 9.95WIDD28 pKa = 3.45ADD30 pKa = 4.03TGEE33 pKa = 4.62EE34 pKa = 4.14YY35 pKa = 11.27GPFPSMLEE43 pKa = 3.74AVQDD47 pKa = 3.56MEE49 pKa = 4.73YY50 pKa = 10.7NAEE53 pKa = 4.02SDD55 pKa = 4.01FEE57 pKa = 4.85PGEE60 pKa = 4.24TVEE63 pKa = 4.23EE64 pKa = 4.26AEE66 pKa = 4.22EE67 pKa = 4.25EE68 pKa = 4.01IGISDD73 pKa = 5.04WIDD76 pKa = 3.59PDD78 pKa = 3.36TGEE81 pKa = 4.7PGEE84 pKa = 4.17EE85 pKa = 3.35AHH87 pKa = 7.07RR88 pKa = 11.84PVEE91 pKa = 4.03
MM1 pKa = 7.33KK2 pKa = 10.1HH3 pKa = 6.93LIPEE7 pKa = 4.57TPPDD11 pKa = 3.76YY12 pKa = 11.24DD13 pKa = 3.14HH14 pKa = 6.79TRR16 pKa = 11.84IIEE19 pKa = 4.29RR20 pKa = 11.84PDD22 pKa = 2.75GCYY25 pKa = 9.95WIDD28 pKa = 3.45ADD30 pKa = 4.03TGEE33 pKa = 4.62EE34 pKa = 4.14YY35 pKa = 11.27GPFPSMLEE43 pKa = 3.74AVQDD47 pKa = 3.56MEE49 pKa = 4.73YY50 pKa = 10.7NAEE53 pKa = 4.02SDD55 pKa = 4.01FEE57 pKa = 4.85PGEE60 pKa = 4.24TVEE63 pKa = 4.23EE64 pKa = 4.26AEE66 pKa = 4.22EE67 pKa = 4.25EE68 pKa = 4.01IGISDD73 pKa = 5.04WIDD76 pKa = 3.59PDD78 pKa = 3.36TGEE81 pKa = 4.7PGEE84 pKa = 4.17EE85 pKa = 3.35AHH87 pKa = 7.07RR88 pKa = 11.84PVEE91 pKa = 4.03
Molecular weight: 10.37 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A125BC38|A0A125BC38_THIDE Cytidylate kinase OS=Thiobacillus denitrificans OX=36861 GN=cmk PE=3 SV=1
MM1 pKa = 7.53FEE3 pKa = 4.63HH4 pKa = 7.02FSDD7 pKa = 4.47ALILADD13 pKa = 3.42AEE15 pKa = 4.36ARR17 pKa = 11.84VSYY20 pKa = 10.19LNPAAEE26 pKa = 4.33RR27 pKa = 11.84LLGLPLNNTRR37 pKa = 11.84GHH39 pKa = 6.78ALDD42 pKa = 5.0KK43 pKa = 11.24VLTLQDD49 pKa = 3.21GLTRR53 pKa = 11.84QSIQIGDD60 pKa = 4.25FPTQHH65 pKa = 6.46SPFSGAFHH73 pKa = 6.74LLVRR77 pKa = 11.84NGGSAIPMQCSVALTRR93 pKa = 11.84TGSGATGGYY102 pKa = 9.19MIVLRR107 pKa = 11.84NASDD111 pKa = 3.7LQQHH115 pKa = 6.12IDD117 pKa = 3.47KK118 pKa = 10.92LVTQSMHH125 pKa = 7.11DD126 pKa = 3.3EE127 pKa = 4.04HH128 pKa = 8.44SRR130 pKa = 11.84LLRR133 pKa = 11.84RR134 pKa = 11.84AEE136 pKa = 3.84LVKK139 pKa = 10.73RR140 pKa = 11.84LWRR143 pKa = 11.84LLQEE147 pKa = 4.52ADD149 pKa = 3.41GGEE152 pKa = 4.23PQAFMYY158 pKa = 10.96LDD160 pKa = 4.2LDD162 pKa = 3.81NFKK165 pKa = 10.93SVNDD169 pKa = 4.11MAGHH173 pKa = 6.89AAGDD177 pKa = 3.53LAIRR181 pKa = 11.84QIAARR186 pKa = 11.84LKK188 pKa = 9.15DD189 pKa = 3.59QVRR192 pKa = 11.84GRR194 pKa = 11.84DD195 pKa = 3.2TLARR199 pKa = 11.84LGGMNSACCWSAVRR213 pKa = 11.84RR214 pKa = 11.84SARR217 pKa = 11.84ASVLSNCIGRR227 pKa = 11.84WNPKK231 pKa = 9.43SCAGTAKK238 pKa = 9.35STGWVSASASPFSKK252 pKa = 9.56RR253 pKa = 11.84TSTARR258 pKa = 11.84TAFWPRR264 pKa = 11.84RR265 pKa = 11.84MRR267 pKa = 11.84RR268 pKa = 11.84AIRR271 pKa = 11.84PSVMAVMGPTYY282 pKa = 10.13RR283 pKa = 11.84KK284 pKa = 9.0SHH286 pKa = 4.52STEE289 pKa = 3.62RR290 pKa = 3.59
MM1 pKa = 7.53FEE3 pKa = 4.63HH4 pKa = 7.02FSDD7 pKa = 4.47ALILADD13 pKa = 3.42AEE15 pKa = 4.36ARR17 pKa = 11.84VSYY20 pKa = 10.19LNPAAEE26 pKa = 4.33RR27 pKa = 11.84LLGLPLNNTRR37 pKa = 11.84GHH39 pKa = 6.78ALDD42 pKa = 5.0KK43 pKa = 11.24VLTLQDD49 pKa = 3.21GLTRR53 pKa = 11.84QSIQIGDD60 pKa = 4.25FPTQHH65 pKa = 6.46SPFSGAFHH73 pKa = 6.74LLVRR77 pKa = 11.84NGGSAIPMQCSVALTRR93 pKa = 11.84TGSGATGGYY102 pKa = 9.19MIVLRR107 pKa = 11.84NASDD111 pKa = 3.7LQQHH115 pKa = 6.12IDD117 pKa = 3.47KK118 pKa = 10.92LVTQSMHH125 pKa = 7.11DD126 pKa = 3.3EE127 pKa = 4.04HH128 pKa = 8.44SRR130 pKa = 11.84LLRR133 pKa = 11.84RR134 pKa = 11.84AEE136 pKa = 3.84LVKK139 pKa = 10.73RR140 pKa = 11.84LWRR143 pKa = 11.84LLQEE147 pKa = 4.52ADD149 pKa = 3.41GGEE152 pKa = 4.23PQAFMYY158 pKa = 10.96LDD160 pKa = 4.2LDD162 pKa = 3.81NFKK165 pKa = 10.93SVNDD169 pKa = 4.11MAGHH173 pKa = 6.89AAGDD177 pKa = 3.53LAIRR181 pKa = 11.84QIAARR186 pKa = 11.84LKK188 pKa = 9.15DD189 pKa = 3.59QVRR192 pKa = 11.84GRR194 pKa = 11.84DD195 pKa = 3.2TLARR199 pKa = 11.84LGGMNSACCWSAVRR213 pKa = 11.84RR214 pKa = 11.84SARR217 pKa = 11.84ASVLSNCIGRR227 pKa = 11.84WNPKK231 pKa = 9.43SCAGTAKK238 pKa = 9.35STGWVSASASPFSKK252 pKa = 9.56RR253 pKa = 11.84TSTARR258 pKa = 11.84TAFWPRR264 pKa = 11.84RR265 pKa = 11.84MRR267 pKa = 11.84RR268 pKa = 11.84AIRR271 pKa = 11.84PSVMAVMGPTYY282 pKa = 10.13RR283 pKa = 11.84KK284 pKa = 9.0SHH286 pKa = 4.52STEE289 pKa = 3.62RR290 pKa = 3.59
Molecular weight: 31.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
919040 |
27 |
3500 |
298.6 |
32.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.323 ± 0.059 | 0.887 ± 0.016 |
5.552 ± 0.034 | 5.549 ± 0.041 |
3.603 ± 0.029 | 7.921 ± 0.043 |
2.395 ± 0.028 | 4.816 ± 0.03 |
3.527 ± 0.042 | 11.109 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.514 ± 0.021 | 2.831 ± 0.027 |
5.023 ± 0.032 | 3.903 ± 0.032 |
6.688 ± 0.046 | 5.064 ± 0.033 |
5.106 ± 0.034 | 7.381 ± 0.037 |
1.386 ± 0.02 | 2.422 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
