
Xingshan nematode virus 4
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Alphanemrhavirus; Xingshan alphanemrhavirus
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins
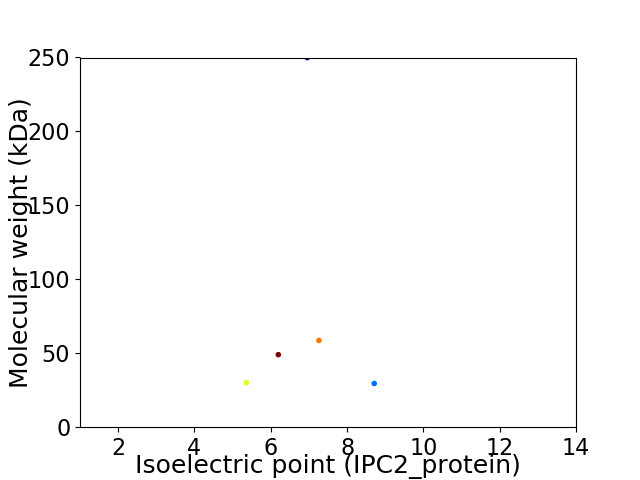
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
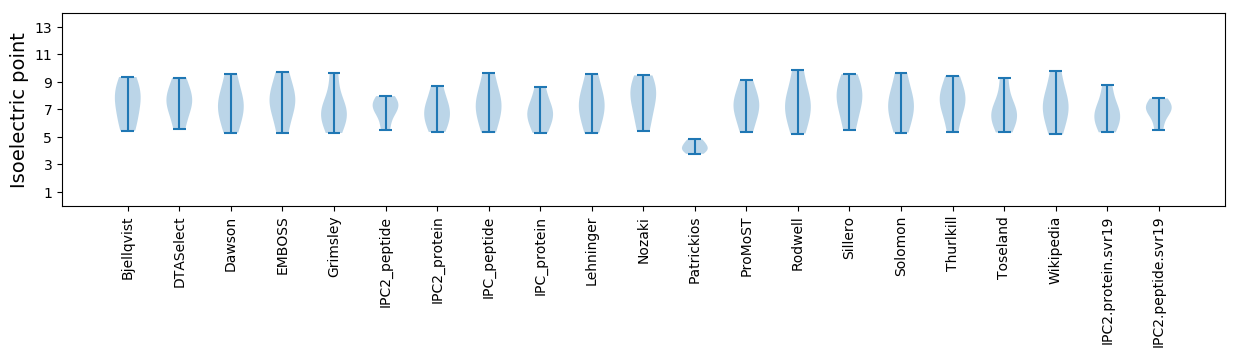
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KN93|A0A1L3KN93_9RHAB GDP polyribonucleotidyltransferase OS=Xingshan nematode virus 4 OX=1923763 PE=4 SV=1
MM1 pKa = 7.34TEE3 pKa = 3.57GRR5 pKa = 11.84LFFEE9 pKa = 4.79PEE11 pKa = 4.12DD12 pKa = 3.65VKK14 pKa = 10.73STNWIPQQANYY25 pKa = 10.69LKK27 pKa = 9.92TDD29 pKa = 3.29LRR31 pKa = 11.84KK32 pKa = 10.26DD33 pKa = 3.55RR34 pKa = 11.84EE35 pKa = 4.42LTGMDD40 pKa = 4.31DD41 pKa = 4.11LEE43 pKa = 4.65SSNFLDD49 pKa = 4.71DD50 pKa = 5.64DD51 pKa = 4.78VYY53 pKa = 11.51SPPPRR58 pKa = 11.84IPFGEE63 pKa = 4.42LSQDD67 pKa = 3.3TLKK70 pKa = 10.11TVQPVEE76 pKa = 4.23NPSGNTSPGFTTYY89 pKa = 10.91KK90 pKa = 8.91FTPIGDD96 pKa = 3.56NPEE99 pKa = 3.86FQRR102 pKa = 11.84RR103 pKa = 11.84LVSEE107 pKa = 4.11TLKK110 pKa = 11.03DD111 pKa = 3.74LEE113 pKa = 4.41KK114 pKa = 10.95LHH116 pKa = 6.86LFTIIHH122 pKa = 5.64EE123 pKa = 4.19QRR125 pKa = 11.84EE126 pKa = 4.26NQSLVFQLKK135 pKa = 9.78RR136 pKa = 11.84KK137 pKa = 7.17TRR139 pKa = 11.84FPSTHH144 pKa = 6.42SGISEE149 pKa = 3.94INPEE153 pKa = 4.1EE154 pKa = 4.22RR155 pKa = 11.84RR156 pKa = 11.84SSVPTDD162 pKa = 3.18QLRR165 pKa = 11.84EE166 pKa = 3.89IVQEE170 pKa = 3.73KK171 pKa = 9.95SRR173 pKa = 11.84IYY175 pKa = 10.59SVQTKK180 pKa = 9.47ARR182 pKa = 11.84KK183 pKa = 9.31IVDD186 pKa = 4.16DD187 pKa = 4.65FPCPVKK193 pKa = 10.47MIGASGSPFKK203 pKa = 11.1INWKK207 pKa = 9.33EE208 pKa = 3.65INVQLEE214 pKa = 4.19QEE216 pKa = 4.27LMAFTEE222 pKa = 4.46EE223 pKa = 4.09EE224 pKa = 4.24LEE226 pKa = 4.14EE227 pKa = 4.35LGKK230 pKa = 10.73LSHH233 pKa = 7.34DD234 pKa = 4.44RR235 pKa = 11.84ILGKK239 pKa = 10.63LLIKK243 pKa = 10.56NHH245 pKa = 6.28LSSPVTKK252 pKa = 10.15LISYY256 pKa = 9.94IYY258 pKa = 10.51FEE260 pKa = 4.3
MM1 pKa = 7.34TEE3 pKa = 3.57GRR5 pKa = 11.84LFFEE9 pKa = 4.79PEE11 pKa = 4.12DD12 pKa = 3.65VKK14 pKa = 10.73STNWIPQQANYY25 pKa = 10.69LKK27 pKa = 9.92TDD29 pKa = 3.29LRR31 pKa = 11.84KK32 pKa = 10.26DD33 pKa = 3.55RR34 pKa = 11.84EE35 pKa = 4.42LTGMDD40 pKa = 4.31DD41 pKa = 4.11LEE43 pKa = 4.65SSNFLDD49 pKa = 4.71DD50 pKa = 5.64DD51 pKa = 4.78VYY53 pKa = 11.51SPPPRR58 pKa = 11.84IPFGEE63 pKa = 4.42LSQDD67 pKa = 3.3TLKK70 pKa = 10.11TVQPVEE76 pKa = 4.23NPSGNTSPGFTTYY89 pKa = 10.91KK90 pKa = 8.91FTPIGDD96 pKa = 3.56NPEE99 pKa = 3.86FQRR102 pKa = 11.84RR103 pKa = 11.84LVSEE107 pKa = 4.11TLKK110 pKa = 11.03DD111 pKa = 3.74LEE113 pKa = 4.41KK114 pKa = 10.95LHH116 pKa = 6.86LFTIIHH122 pKa = 5.64EE123 pKa = 4.19QRR125 pKa = 11.84EE126 pKa = 4.26NQSLVFQLKK135 pKa = 9.78RR136 pKa = 11.84KK137 pKa = 7.17TRR139 pKa = 11.84FPSTHH144 pKa = 6.42SGISEE149 pKa = 3.94INPEE153 pKa = 4.1EE154 pKa = 4.22RR155 pKa = 11.84RR156 pKa = 11.84SSVPTDD162 pKa = 3.18QLRR165 pKa = 11.84EE166 pKa = 3.89IVQEE170 pKa = 3.73KK171 pKa = 9.95SRR173 pKa = 11.84IYY175 pKa = 10.59SVQTKK180 pKa = 9.47ARR182 pKa = 11.84KK183 pKa = 9.31IVDD186 pKa = 4.16DD187 pKa = 4.65FPCPVKK193 pKa = 10.47MIGASGSPFKK203 pKa = 11.1INWKK207 pKa = 9.33EE208 pKa = 3.65INVQLEE214 pKa = 4.19QEE216 pKa = 4.27LMAFTEE222 pKa = 4.46EE223 pKa = 4.09EE224 pKa = 4.24LEE226 pKa = 4.14EE227 pKa = 4.35LGKK230 pKa = 10.73LSHH233 pKa = 7.34DD234 pKa = 4.44RR235 pKa = 11.84ILGKK239 pKa = 10.63LLIKK243 pKa = 10.56NHH245 pKa = 6.28LSSPVTKK252 pKa = 10.15LISYY256 pKa = 9.94IYY258 pKa = 10.51FEE260 pKa = 4.3
Molecular weight: 30.08 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KNG7|A0A1L3KNG7_9RHAB Uncharacterized protein OS=Xingshan nematode virus 4 OX=1923763 PE=4 SV=1
MM1 pKa = 7.71NYY3 pKa = 10.49LPILRR8 pKa = 11.84RR9 pKa = 11.84LNKK12 pKa = 10.16NGNRR16 pKa = 11.84NEE18 pKa = 3.92NRR20 pKa = 11.84NDD22 pKa = 3.24NTKK25 pKa = 10.56NPSNLGEE32 pKa = 4.1NFKK35 pKa = 10.55PKK37 pKa = 9.95KK38 pKa = 10.22DD39 pKa = 3.54QTFLTDD45 pKa = 4.02FPKK48 pKa = 11.27GEE50 pKa = 4.13FKK52 pKa = 11.0NFNSIQTSSRR62 pKa = 11.84SPQLYY67 pKa = 8.94FDD69 pKa = 5.21KK70 pKa = 10.74IEE72 pKa = 4.18KK73 pKa = 10.1PIVEE77 pKa = 4.7LSCGPKK83 pKa = 10.02FSKK86 pKa = 10.5EE87 pKa = 4.17YY88 pKa = 9.94IEE90 pKa = 4.41KK91 pKa = 10.11VHH93 pKa = 5.48YY94 pKa = 9.02TLNLEE99 pKa = 4.24VNAEE103 pKa = 4.08VTILTTKK110 pKa = 10.64RR111 pKa = 11.84LTIQEE116 pKa = 4.25IAGHH120 pKa = 5.48LLEE123 pKa = 4.59HH124 pKa = 7.05RR125 pKa = 11.84EE126 pKa = 4.0RR127 pKa = 11.84FVGDD131 pKa = 3.1LRR133 pKa = 11.84IYY135 pKa = 9.62PIYY138 pKa = 9.94HH139 pKa = 5.54ITMLLCLHH147 pKa = 6.16NCKK150 pKa = 10.06HH151 pKa = 6.0QDD153 pKa = 2.83ITRR156 pKa = 11.84MEE158 pKa = 4.6FKK160 pKa = 10.38HH161 pKa = 6.1QYY163 pKa = 7.67SNHH166 pKa = 5.84MEE168 pKa = 4.02TVLQINTKK176 pKa = 9.45FSKK179 pKa = 11.06DD180 pKa = 3.32NVISKK185 pKa = 10.56NGVAYY190 pKa = 9.88RR191 pKa = 11.84VEE193 pKa = 4.1WTNLGIDD200 pKa = 4.09YY201 pKa = 10.19KK202 pKa = 9.63LHH204 pKa = 5.0WMFRR208 pKa = 11.84TFLKK212 pKa = 9.49KK213 pKa = 7.81TTVKK217 pKa = 10.58GISPTYY223 pKa = 9.45QQFPLLPYY231 pKa = 10.11FLEE234 pKa = 4.61ASALSVCLKK243 pKa = 7.8EE244 pKa = 4.17TNLIITQQ251 pKa = 3.63
MM1 pKa = 7.71NYY3 pKa = 10.49LPILRR8 pKa = 11.84RR9 pKa = 11.84LNKK12 pKa = 10.16NGNRR16 pKa = 11.84NEE18 pKa = 3.92NRR20 pKa = 11.84NDD22 pKa = 3.24NTKK25 pKa = 10.56NPSNLGEE32 pKa = 4.1NFKK35 pKa = 10.55PKK37 pKa = 9.95KK38 pKa = 10.22DD39 pKa = 3.54QTFLTDD45 pKa = 4.02FPKK48 pKa = 11.27GEE50 pKa = 4.13FKK52 pKa = 11.0NFNSIQTSSRR62 pKa = 11.84SPQLYY67 pKa = 8.94FDD69 pKa = 5.21KK70 pKa = 10.74IEE72 pKa = 4.18KK73 pKa = 10.1PIVEE77 pKa = 4.7LSCGPKK83 pKa = 10.02FSKK86 pKa = 10.5EE87 pKa = 4.17YY88 pKa = 9.94IEE90 pKa = 4.41KK91 pKa = 10.11VHH93 pKa = 5.48YY94 pKa = 9.02TLNLEE99 pKa = 4.24VNAEE103 pKa = 4.08VTILTTKK110 pKa = 10.64RR111 pKa = 11.84LTIQEE116 pKa = 4.25IAGHH120 pKa = 5.48LLEE123 pKa = 4.59HH124 pKa = 7.05RR125 pKa = 11.84EE126 pKa = 4.0RR127 pKa = 11.84FVGDD131 pKa = 3.1LRR133 pKa = 11.84IYY135 pKa = 9.62PIYY138 pKa = 9.94HH139 pKa = 5.54ITMLLCLHH147 pKa = 6.16NCKK150 pKa = 10.06HH151 pKa = 6.0QDD153 pKa = 2.83ITRR156 pKa = 11.84MEE158 pKa = 4.6FKK160 pKa = 10.38HH161 pKa = 6.1QYY163 pKa = 7.67SNHH166 pKa = 5.84MEE168 pKa = 4.02TVLQINTKK176 pKa = 9.45FSKK179 pKa = 11.06DD180 pKa = 3.32NVISKK185 pKa = 10.56NGVAYY190 pKa = 9.88RR191 pKa = 11.84VEE193 pKa = 4.1WTNLGIDD200 pKa = 4.09YY201 pKa = 10.19KK202 pKa = 9.63LHH204 pKa = 5.0WMFRR208 pKa = 11.84TFLKK212 pKa = 9.49KK213 pKa = 7.81TTVKK217 pKa = 10.58GISPTYY223 pKa = 9.45QQFPLLPYY231 pKa = 10.11FLEE234 pKa = 4.61ASALSVCLKK243 pKa = 7.8EE244 pKa = 4.17TNLIITQQ251 pKa = 3.63
Molecular weight: 29.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3625 |
251 |
2169 |
725.0 |
83.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.614 ± 0.582 | 2.207 ± 0.332 |
4.579 ± 0.342 | 6.345 ± 0.544 |
5.241 ± 0.228 | 5.103 ± 0.259 |
3.09 ± 0.23 | 7.531 ± 0.502 |
6.179 ± 0.786 | 11.752 ± 0.686 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.848 ± 0.318 | 5.103 ± 0.681 |
4.69 ± 0.312 | 3.476 ± 0.328 |
4.993 ± 0.462 | 8.303 ± 0.651 |
5.71 ± 0.735 | 4.662 ± 0.241 |
1.628 ± 0.343 | 3.945 ± 0.224 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
