
Salvia hispanica RNA virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Amalgaviridae; unclassified Amalgaviridae
Average proteome isoelectric point is 6.86
Get precalculated fractions of proteins
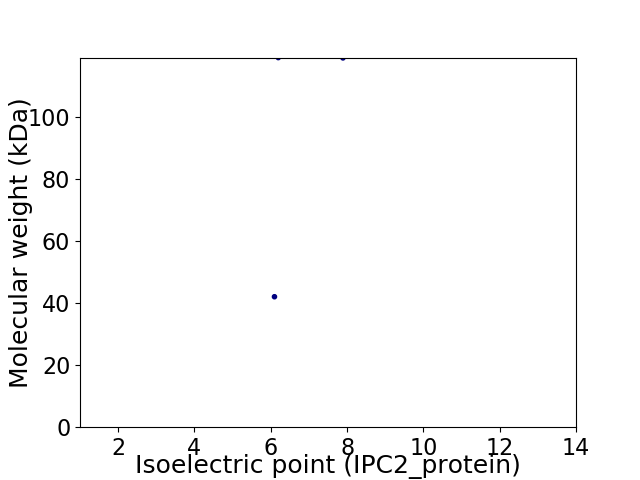
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
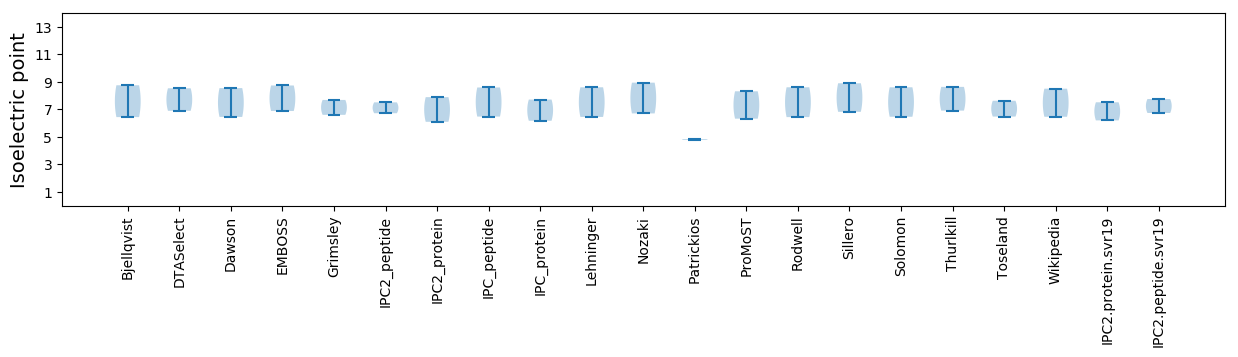
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A386RWS8|A0A386RWS8_9VIRU Coat protein OS=Salvia hispanica RNA virus 1 OX=2419805 PE=4 SV=1
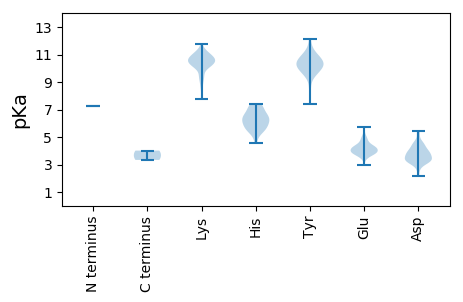
MM1 pKa = 7.26AAEE4 pKa = 4.23APRR7 pKa = 11.84RR8 pKa = 11.84VLAVKK13 pKa = 8.75TPPNYY18 pKa = 10.81DD19 pKa = 3.25EE20 pKa = 4.96MLTNTLAPLAAEE32 pKa = 4.86AFPVHH37 pKa = 5.42TWTRR41 pKa = 11.84AALLANYY48 pKa = 10.23LDD50 pKa = 3.19IKK52 pKa = 10.87RR53 pKa = 11.84FIDD56 pKa = 3.45HH57 pKa = 6.32VKK59 pKa = 10.56VISGEE64 pKa = 3.83NDD66 pKa = 3.24PVIRR70 pKa = 11.84TAMIRR75 pKa = 11.84KK76 pKa = 9.24GNTSAAWNTQGTCTASQMFKK96 pKa = 10.09FCAWLKK102 pKa = 10.57SPEE105 pKa = 4.07GNKK108 pKa = 10.22FITTEE113 pKa = 3.47RR114 pKa = 11.84RR115 pKa = 11.84KK116 pKa = 10.59RR117 pKa = 11.84NIEE120 pKa = 3.48KK121 pKa = 10.38RR122 pKa = 11.84AFEE125 pKa = 4.27GQQFTDD131 pKa = 3.35SSANAAMEE139 pKa = 4.05AQITEE144 pKa = 3.87HH145 pKa = 6.6AMLVKK150 pKa = 10.58KK151 pKa = 10.66SRR153 pKa = 11.84ATTEE157 pKa = 3.73EE158 pKa = 3.85TLLQLRR164 pKa = 11.84RR165 pKa = 11.84EE166 pKa = 4.26IARR169 pKa = 11.84TQQRR173 pKa = 11.84GEE175 pKa = 3.82ADD177 pKa = 3.26IKK179 pKa = 11.24ALEE182 pKa = 4.3ADD184 pKa = 4.72FAPGSAYY191 pKa = 10.79VPMDD195 pKa = 3.98DD196 pKa = 4.01QDD198 pKa = 3.91LGRR201 pKa = 11.84ACHH204 pKa = 5.85QLYY207 pKa = 10.14LAQCQRR213 pKa = 11.84EE214 pKa = 4.17DD215 pKa = 4.5ADD217 pKa = 4.43PEE219 pKa = 4.33DD220 pKa = 3.81LTEE223 pKa = 5.74EE224 pKa = 3.69IMADD228 pKa = 3.11IKK230 pKa = 11.03ATFGSEE236 pKa = 2.94ATARR240 pKa = 11.84HH241 pKa = 5.49RR242 pKa = 11.84AAFIAEE248 pKa = 3.83GHH250 pKa = 6.09RR251 pKa = 11.84RR252 pKa = 11.84QDD254 pKa = 3.4LLAWTEE260 pKa = 3.9AKK262 pKa = 10.12VQQLAGLGDD271 pKa = 4.53LKK273 pKa = 10.93RR274 pKa = 11.84ADD276 pKa = 3.64TFRR279 pKa = 11.84VLVEE283 pKa = 4.09GMGGEE288 pKa = 4.75LSAQEE293 pKa = 3.78RR294 pKa = 11.84AAAEE298 pKa = 4.03ARR300 pKa = 11.84AVAAGGLGPNRR311 pKa = 11.84RR312 pKa = 11.84RR313 pKa = 11.84RR314 pKa = 11.84RR315 pKa = 11.84ADD317 pKa = 3.07VDD319 pKa = 3.39PQNIIDD325 pKa = 3.62HH326 pKa = 5.93PRR328 pKa = 11.84RR329 pKa = 11.84RR330 pKa = 11.84GNQASVLQVEE340 pKa = 5.07APPPPVVQGGDD351 pKa = 3.0AGSRR355 pKa = 11.84AGSPSNPPTDD365 pKa = 3.07IRR367 pKa = 11.84IASPRR372 pKa = 11.84NEE374 pKa = 4.27GGGEE378 pKa = 3.96NPDD381 pKa = 3.36VQEE384 pKa = 4.01
MM1 pKa = 7.26AAEE4 pKa = 4.23APRR7 pKa = 11.84RR8 pKa = 11.84VLAVKK13 pKa = 8.75TPPNYY18 pKa = 10.81DD19 pKa = 3.25EE20 pKa = 4.96MLTNTLAPLAAEE32 pKa = 4.86AFPVHH37 pKa = 5.42TWTRR41 pKa = 11.84AALLANYY48 pKa = 10.23LDD50 pKa = 3.19IKK52 pKa = 10.87RR53 pKa = 11.84FIDD56 pKa = 3.45HH57 pKa = 6.32VKK59 pKa = 10.56VISGEE64 pKa = 3.83NDD66 pKa = 3.24PVIRR70 pKa = 11.84TAMIRR75 pKa = 11.84KK76 pKa = 9.24GNTSAAWNTQGTCTASQMFKK96 pKa = 10.09FCAWLKK102 pKa = 10.57SPEE105 pKa = 4.07GNKK108 pKa = 10.22FITTEE113 pKa = 3.47RR114 pKa = 11.84RR115 pKa = 11.84KK116 pKa = 10.59RR117 pKa = 11.84NIEE120 pKa = 3.48KK121 pKa = 10.38RR122 pKa = 11.84AFEE125 pKa = 4.27GQQFTDD131 pKa = 3.35SSANAAMEE139 pKa = 4.05AQITEE144 pKa = 3.87HH145 pKa = 6.6AMLVKK150 pKa = 10.58KK151 pKa = 10.66SRR153 pKa = 11.84ATTEE157 pKa = 3.73EE158 pKa = 3.85TLLQLRR164 pKa = 11.84RR165 pKa = 11.84EE166 pKa = 4.26IARR169 pKa = 11.84TQQRR173 pKa = 11.84GEE175 pKa = 3.82ADD177 pKa = 3.26IKK179 pKa = 11.24ALEE182 pKa = 4.3ADD184 pKa = 4.72FAPGSAYY191 pKa = 10.79VPMDD195 pKa = 3.98DD196 pKa = 4.01QDD198 pKa = 3.91LGRR201 pKa = 11.84ACHH204 pKa = 5.85QLYY207 pKa = 10.14LAQCQRR213 pKa = 11.84EE214 pKa = 4.17DD215 pKa = 4.5ADD217 pKa = 4.43PEE219 pKa = 4.33DD220 pKa = 3.81LTEE223 pKa = 5.74EE224 pKa = 3.69IMADD228 pKa = 3.11IKK230 pKa = 11.03ATFGSEE236 pKa = 2.94ATARR240 pKa = 11.84HH241 pKa = 5.49RR242 pKa = 11.84AAFIAEE248 pKa = 3.83GHH250 pKa = 6.09RR251 pKa = 11.84RR252 pKa = 11.84QDD254 pKa = 3.4LLAWTEE260 pKa = 3.9AKK262 pKa = 10.12VQQLAGLGDD271 pKa = 4.53LKK273 pKa = 10.93RR274 pKa = 11.84ADD276 pKa = 3.64TFRR279 pKa = 11.84VLVEE283 pKa = 4.09GMGGEE288 pKa = 4.75LSAQEE293 pKa = 3.78RR294 pKa = 11.84AAAEE298 pKa = 4.03ARR300 pKa = 11.84AVAAGGLGPNRR311 pKa = 11.84RR312 pKa = 11.84RR313 pKa = 11.84RR314 pKa = 11.84RR315 pKa = 11.84ADD317 pKa = 3.07VDD319 pKa = 3.39PQNIIDD325 pKa = 3.62HH326 pKa = 5.93PRR328 pKa = 11.84RR329 pKa = 11.84RR330 pKa = 11.84GNQASVLQVEE340 pKa = 5.07APPPPVVQGGDD351 pKa = 3.0AGSRR355 pKa = 11.84AGSPSNPPTDD365 pKa = 3.07IRR367 pKa = 11.84IASPRR372 pKa = 11.84NEE374 pKa = 4.27GGGEE378 pKa = 3.96NPDD381 pKa = 3.36VQEE384 pKa = 4.01
Molecular weight: 42.11 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A386RWS8|A0A386RWS8_9VIRU Coat protein OS=Salvia hispanica RNA virus 1 OX=2419805 PE=4 SV=1
MM1 pKa = 7.26AAEE4 pKa = 4.23APRR7 pKa = 11.84RR8 pKa = 11.84VLAVKK13 pKa = 8.75TPPNYY18 pKa = 10.81DD19 pKa = 3.25EE20 pKa = 4.96MLTNTLAPLAAEE32 pKa = 4.86AFPVHH37 pKa = 5.42TWTRR41 pKa = 11.84AALLANYY48 pKa = 10.23LDD50 pKa = 3.19IKK52 pKa = 10.87RR53 pKa = 11.84FIDD56 pKa = 3.45HH57 pKa = 6.32VKK59 pKa = 10.56VISGEE64 pKa = 3.83NDD66 pKa = 3.24PVIRR70 pKa = 11.84TAMIRR75 pKa = 11.84KK76 pKa = 9.24GNTSAAWNTQGTCTASQMFKK96 pKa = 10.09FCAWLKK102 pKa = 10.57SPEE105 pKa = 4.07GNKK108 pKa = 10.22FITTEE113 pKa = 3.47RR114 pKa = 11.84RR115 pKa = 11.84KK116 pKa = 10.59RR117 pKa = 11.84NIEE120 pKa = 3.48KK121 pKa = 10.38RR122 pKa = 11.84AFEE125 pKa = 4.27GQQFTDD131 pKa = 3.35SSANAAMEE139 pKa = 4.05AQITEE144 pKa = 3.87HH145 pKa = 6.6AMLVKK150 pKa = 10.58KK151 pKa = 10.66SRR153 pKa = 11.84ATTEE157 pKa = 3.73EE158 pKa = 3.85TLLQLRR164 pKa = 11.84RR165 pKa = 11.84EE166 pKa = 4.26IARR169 pKa = 11.84TQQRR173 pKa = 11.84GEE175 pKa = 3.82ADD177 pKa = 3.26IKK179 pKa = 11.24ALEE182 pKa = 4.3ADD184 pKa = 4.72FAPGSAYY191 pKa = 10.79VPMDD195 pKa = 3.98DD196 pKa = 4.01QDD198 pKa = 3.91LGRR201 pKa = 11.84ACHH204 pKa = 5.85QLYY207 pKa = 10.14LAQCQRR213 pKa = 11.84EE214 pKa = 4.17DD215 pKa = 4.5ADD217 pKa = 4.43PEE219 pKa = 4.33DD220 pKa = 3.81LTEE223 pKa = 5.74EE224 pKa = 3.69IMADD228 pKa = 3.11IKK230 pKa = 11.03ATFGSEE236 pKa = 2.94ATARR240 pKa = 11.84HH241 pKa = 5.49RR242 pKa = 11.84AAFIAEE248 pKa = 3.83GHH250 pKa = 6.09RR251 pKa = 11.84RR252 pKa = 11.84QDD254 pKa = 3.4LLAWTEE260 pKa = 3.9AKK262 pKa = 10.12VQQLAGLGDD271 pKa = 4.53LKK273 pKa = 11.02RR274 pKa = 11.84ADD276 pKa = 3.77TFVSSWKK283 pKa = 9.5EE284 pKa = 3.48WAEE287 pKa = 3.92SFLLKK292 pKa = 10.37RR293 pKa = 11.84EE294 pKa = 3.77PRR296 pKa = 11.84LRR298 pKa = 11.84RR299 pKa = 11.84EE300 pKa = 4.12LLQQVVWGPIAAGGGLTSTHH320 pKa = 7.14RR321 pKa = 11.84ISSIIPADD329 pKa = 3.74VVTKK333 pKa = 7.78PQCSKK338 pKa = 10.74SKK340 pKa = 10.24RR341 pKa = 11.84RR342 pKa = 11.84PPLSSKK348 pKa = 10.89VEE350 pKa = 4.16TLDD353 pKa = 3.78LAPGLLVTHH362 pKa = 6.05QRR364 pKa = 11.84TSVLRR369 pKa = 11.84VPAMKK374 pKa = 10.52GGGRR378 pKa = 11.84IPTSRR383 pKa = 11.84SKK385 pKa = 11.24FEE387 pKa = 3.94AKK389 pKa = 9.08VRR391 pKa = 11.84RR392 pKa = 11.84VIGGGEE398 pKa = 3.7MRR400 pKa = 11.84NWAKK404 pKa = 10.74DD405 pKa = 3.29SNMYY409 pKa = 10.39RR410 pKa = 11.84GGGNYY415 pKa = 9.86SDD417 pKa = 4.29ALKK420 pKa = 10.95LLATARR426 pKa = 11.84TDD428 pKa = 3.09IPGSFLHH435 pKa = 6.77KK436 pKa = 10.32HH437 pKa = 4.58FTVNNARR444 pKa = 11.84SFLKK448 pKa = 10.41LPCGLPVPVGPEE460 pKa = 3.83SVRR463 pKa = 11.84MKK465 pKa = 10.82NFNEE469 pKa = 4.09EE470 pKa = 3.78ATAGPSFRR478 pKa = 11.84AFGIYY483 pKa = 9.49RR484 pKa = 11.84KK485 pKa = 9.9KK486 pKa = 10.53GLKK489 pKa = 10.11AEE491 pKa = 4.48LEE493 pKa = 4.16ATVWKK498 pKa = 10.29SLHH501 pKa = 6.57AYY503 pKa = 10.31AEE505 pKa = 4.55GGDD508 pKa = 4.21AEE510 pKa = 4.49SCLPFIAARR519 pKa = 11.84VGYY522 pKa = 9.64RR523 pKa = 11.84SKK525 pKa = 11.42LLTLEE530 pKa = 4.19KK531 pKa = 10.56AFSKK535 pKa = 9.52LTSGEE540 pKa = 4.11CLGRR544 pKa = 11.84CVMMLDD550 pKa = 4.28AFEE553 pKa = 4.2QTYY556 pKa = 11.16SSALYY561 pKa = 10.28NVISGITHH569 pKa = 7.21RR570 pKa = 11.84GRR572 pKa = 11.84HH573 pKa = 5.11NPGSSFRR580 pKa = 11.84NTTVRR585 pKa = 11.84ASSDD589 pKa = 2.37WGMLFEE595 pKa = 4.36EE596 pKa = 4.79VKK598 pKa = 10.53KK599 pKa = 10.82ASAVIEE605 pKa = 4.2LDD607 pKa = 2.83WKK609 pKa = 11.04KK610 pKa = 10.65FDD612 pKa = 4.63RR613 pKa = 11.84DD614 pKa = 3.58RR615 pKa = 11.84PSDD618 pKa = 4.27DD619 pKa = 3.18IYY621 pKa = 11.37FVIDD625 pKa = 4.45IILSCFQPKK634 pKa = 9.97NGYY637 pKa = 7.39EE638 pKa = 3.82ARR640 pKa = 11.84LLEE643 pKa = 3.9AHH645 pKa = 7.23GIMLRR650 pKa = 11.84RR651 pKa = 11.84ALVEE655 pKa = 4.2RR656 pKa = 11.84PFITDD661 pKa = 3.19DD662 pKa = 3.41GGIFTIEE669 pKa = 3.92GMVPSGSLWTGWLDD683 pKa = 3.45TALNTLYY690 pKa = 10.31IKK692 pKa = 10.71AVLFHH697 pKa = 6.76LGFRR701 pKa = 11.84EE702 pKa = 3.93SDD704 pKa = 3.46ASPKK708 pKa = 10.43CAGDD712 pKa = 4.07DD713 pKa = 3.55NLTLIFKK720 pKa = 9.43EE721 pKa = 4.1ATDD724 pKa = 3.75MQLMEE729 pKa = 4.72VKK731 pKa = 10.43RR732 pKa = 11.84LLNEE736 pKa = 4.15WFRR739 pKa = 11.84AGIDD743 pKa = 3.89DD744 pKa = 4.2EE745 pKa = 5.68DD746 pKa = 5.36FMVHH750 pKa = 6.31HH751 pKa = 7.19PPYY754 pKa = 10.05HH755 pKa = 5.67VEE757 pKa = 3.55RR758 pKa = 11.84RR759 pKa = 11.84QAVFPPGTDD768 pKa = 3.28LSKK771 pKa = 10.65GTSKK775 pKa = 11.04LLDD778 pKa = 3.68LAQWVPFEE786 pKa = 5.12DD787 pKa = 4.24PMTIDD792 pKa = 3.27EE793 pKa = 4.82SKK795 pKa = 10.96GLSHH799 pKa = 6.55RR800 pKa = 11.84WKK802 pKa = 10.84YY803 pKa = 10.52IFEE806 pKa = 4.46NKK808 pKa = 9.54PKK810 pKa = 10.38FLSCYY815 pKa = 8.45WDD817 pKa = 3.59EE818 pKa = 4.44NNNPIRR824 pKa = 11.84PAYY827 pKa = 9.63INLEE831 pKa = 3.91KK832 pKa = 10.85LLWPEE837 pKa = 5.22GIHH840 pKa = 5.58EE841 pKa = 4.5TIEE844 pKa = 4.41DD845 pKa = 3.93YY846 pKa = 10.87EE847 pKa = 4.18AALIGMVVDD856 pKa = 4.66NPFNHH861 pKa = 6.71HH862 pKa = 5.73NVNHH866 pKa = 6.29LMHH869 pKa = 7.4RR870 pKa = 11.84YY871 pKa = 10.01CIVQQVKK878 pKa = 10.09RR879 pKa = 11.84MMITGIKK886 pKa = 10.32AEE888 pKa = 4.58DD889 pKa = 3.45VLDD892 pKa = 4.15FCRR895 pKa = 11.84FKK897 pKa = 10.99DD898 pKa = 3.82DD899 pKa = 3.82KK900 pKa = 11.74GEE902 pKa = 4.15GVPFPMVAQWRR913 pKa = 11.84RR914 pKa = 11.84VNGWVDD920 pKa = 3.39MEE922 pKa = 4.62KK923 pKa = 10.88LPFVDD928 pKa = 5.42RR929 pKa = 11.84YY930 pKa = 9.53IAQFRR935 pKa = 11.84DD936 pKa = 3.8FVTGVTSLYY945 pKa = 10.58SRR947 pKa = 11.84SPTGGLDD954 pKa = 2.6SWRR957 pKa = 11.84FMDD960 pKa = 5.26IIRR963 pKa = 11.84GVGNLNDD970 pKa = 3.69AQFGNDD976 pKa = 2.84MVDD979 pKa = 2.16WVTFLKK985 pKa = 10.56GHH987 pKa = 7.31PLTKK991 pKa = 10.16YY992 pKa = 10.1LKK994 pKa = 7.9PTKK997 pKa = 10.16GSRR1000 pKa = 11.84KK1001 pKa = 9.32RR1002 pKa = 11.84KK1003 pKa = 8.52LHH1005 pKa = 6.36SDD1007 pKa = 3.64PEE1009 pKa = 4.36EE1010 pKa = 3.9EE1011 pKa = 3.76ALTRR1015 pKa = 11.84FKK1017 pKa = 11.2VLNHH1021 pKa = 6.07YY1022 pKa = 10.52LDD1024 pKa = 4.08PTWKK1028 pKa = 10.42KK1029 pKa = 11.38KK1030 pKa = 10.17MFDD1033 pKa = 4.05DD1034 pKa = 3.81VDD1036 pKa = 5.22SYY1038 pKa = 12.15ALWISDD1044 pKa = 3.69LLRR1047 pKa = 11.84NKK1049 pKa = 10.47GPTSS1053 pKa = 3.34
MM1 pKa = 7.26AAEE4 pKa = 4.23APRR7 pKa = 11.84RR8 pKa = 11.84VLAVKK13 pKa = 8.75TPPNYY18 pKa = 10.81DD19 pKa = 3.25EE20 pKa = 4.96MLTNTLAPLAAEE32 pKa = 4.86AFPVHH37 pKa = 5.42TWTRR41 pKa = 11.84AALLANYY48 pKa = 10.23LDD50 pKa = 3.19IKK52 pKa = 10.87RR53 pKa = 11.84FIDD56 pKa = 3.45HH57 pKa = 6.32VKK59 pKa = 10.56VISGEE64 pKa = 3.83NDD66 pKa = 3.24PVIRR70 pKa = 11.84TAMIRR75 pKa = 11.84KK76 pKa = 9.24GNTSAAWNTQGTCTASQMFKK96 pKa = 10.09FCAWLKK102 pKa = 10.57SPEE105 pKa = 4.07GNKK108 pKa = 10.22FITTEE113 pKa = 3.47RR114 pKa = 11.84RR115 pKa = 11.84KK116 pKa = 10.59RR117 pKa = 11.84NIEE120 pKa = 3.48KK121 pKa = 10.38RR122 pKa = 11.84AFEE125 pKa = 4.27GQQFTDD131 pKa = 3.35SSANAAMEE139 pKa = 4.05AQITEE144 pKa = 3.87HH145 pKa = 6.6AMLVKK150 pKa = 10.58KK151 pKa = 10.66SRR153 pKa = 11.84ATTEE157 pKa = 3.73EE158 pKa = 3.85TLLQLRR164 pKa = 11.84RR165 pKa = 11.84EE166 pKa = 4.26IARR169 pKa = 11.84TQQRR173 pKa = 11.84GEE175 pKa = 3.82ADD177 pKa = 3.26IKK179 pKa = 11.24ALEE182 pKa = 4.3ADD184 pKa = 4.72FAPGSAYY191 pKa = 10.79VPMDD195 pKa = 3.98DD196 pKa = 4.01QDD198 pKa = 3.91LGRR201 pKa = 11.84ACHH204 pKa = 5.85QLYY207 pKa = 10.14LAQCQRR213 pKa = 11.84EE214 pKa = 4.17DD215 pKa = 4.5ADD217 pKa = 4.43PEE219 pKa = 4.33DD220 pKa = 3.81LTEE223 pKa = 5.74EE224 pKa = 3.69IMADD228 pKa = 3.11IKK230 pKa = 11.03ATFGSEE236 pKa = 2.94ATARR240 pKa = 11.84HH241 pKa = 5.49RR242 pKa = 11.84AAFIAEE248 pKa = 3.83GHH250 pKa = 6.09RR251 pKa = 11.84RR252 pKa = 11.84QDD254 pKa = 3.4LLAWTEE260 pKa = 3.9AKK262 pKa = 10.12VQQLAGLGDD271 pKa = 4.53LKK273 pKa = 11.02RR274 pKa = 11.84ADD276 pKa = 3.77TFVSSWKK283 pKa = 9.5EE284 pKa = 3.48WAEE287 pKa = 3.92SFLLKK292 pKa = 10.37RR293 pKa = 11.84EE294 pKa = 3.77PRR296 pKa = 11.84LRR298 pKa = 11.84RR299 pKa = 11.84EE300 pKa = 4.12LLQQVVWGPIAAGGGLTSTHH320 pKa = 7.14RR321 pKa = 11.84ISSIIPADD329 pKa = 3.74VVTKK333 pKa = 7.78PQCSKK338 pKa = 10.74SKK340 pKa = 10.24RR341 pKa = 11.84RR342 pKa = 11.84PPLSSKK348 pKa = 10.89VEE350 pKa = 4.16TLDD353 pKa = 3.78LAPGLLVTHH362 pKa = 6.05QRR364 pKa = 11.84TSVLRR369 pKa = 11.84VPAMKK374 pKa = 10.52GGGRR378 pKa = 11.84IPTSRR383 pKa = 11.84SKK385 pKa = 11.24FEE387 pKa = 3.94AKK389 pKa = 9.08VRR391 pKa = 11.84RR392 pKa = 11.84VIGGGEE398 pKa = 3.7MRR400 pKa = 11.84NWAKK404 pKa = 10.74DD405 pKa = 3.29SNMYY409 pKa = 10.39RR410 pKa = 11.84GGGNYY415 pKa = 9.86SDD417 pKa = 4.29ALKK420 pKa = 10.95LLATARR426 pKa = 11.84TDD428 pKa = 3.09IPGSFLHH435 pKa = 6.77KK436 pKa = 10.32HH437 pKa = 4.58FTVNNARR444 pKa = 11.84SFLKK448 pKa = 10.41LPCGLPVPVGPEE460 pKa = 3.83SVRR463 pKa = 11.84MKK465 pKa = 10.82NFNEE469 pKa = 4.09EE470 pKa = 3.78ATAGPSFRR478 pKa = 11.84AFGIYY483 pKa = 9.49RR484 pKa = 11.84KK485 pKa = 9.9KK486 pKa = 10.53GLKK489 pKa = 10.11AEE491 pKa = 4.48LEE493 pKa = 4.16ATVWKK498 pKa = 10.29SLHH501 pKa = 6.57AYY503 pKa = 10.31AEE505 pKa = 4.55GGDD508 pKa = 4.21AEE510 pKa = 4.49SCLPFIAARR519 pKa = 11.84VGYY522 pKa = 9.64RR523 pKa = 11.84SKK525 pKa = 11.42LLTLEE530 pKa = 4.19KK531 pKa = 10.56AFSKK535 pKa = 9.52LTSGEE540 pKa = 4.11CLGRR544 pKa = 11.84CVMMLDD550 pKa = 4.28AFEE553 pKa = 4.2QTYY556 pKa = 11.16SSALYY561 pKa = 10.28NVISGITHH569 pKa = 7.21RR570 pKa = 11.84GRR572 pKa = 11.84HH573 pKa = 5.11NPGSSFRR580 pKa = 11.84NTTVRR585 pKa = 11.84ASSDD589 pKa = 2.37WGMLFEE595 pKa = 4.36EE596 pKa = 4.79VKK598 pKa = 10.53KK599 pKa = 10.82ASAVIEE605 pKa = 4.2LDD607 pKa = 2.83WKK609 pKa = 11.04KK610 pKa = 10.65FDD612 pKa = 4.63RR613 pKa = 11.84DD614 pKa = 3.58RR615 pKa = 11.84PSDD618 pKa = 4.27DD619 pKa = 3.18IYY621 pKa = 11.37FVIDD625 pKa = 4.45IILSCFQPKK634 pKa = 9.97NGYY637 pKa = 7.39EE638 pKa = 3.82ARR640 pKa = 11.84LLEE643 pKa = 3.9AHH645 pKa = 7.23GIMLRR650 pKa = 11.84RR651 pKa = 11.84ALVEE655 pKa = 4.2RR656 pKa = 11.84PFITDD661 pKa = 3.19DD662 pKa = 3.41GGIFTIEE669 pKa = 3.92GMVPSGSLWTGWLDD683 pKa = 3.45TALNTLYY690 pKa = 10.31IKK692 pKa = 10.71AVLFHH697 pKa = 6.76LGFRR701 pKa = 11.84EE702 pKa = 3.93SDD704 pKa = 3.46ASPKK708 pKa = 10.43CAGDD712 pKa = 4.07DD713 pKa = 3.55NLTLIFKK720 pKa = 9.43EE721 pKa = 4.1ATDD724 pKa = 3.75MQLMEE729 pKa = 4.72VKK731 pKa = 10.43RR732 pKa = 11.84LLNEE736 pKa = 4.15WFRR739 pKa = 11.84AGIDD743 pKa = 3.89DD744 pKa = 4.2EE745 pKa = 5.68DD746 pKa = 5.36FMVHH750 pKa = 6.31HH751 pKa = 7.19PPYY754 pKa = 10.05HH755 pKa = 5.67VEE757 pKa = 3.55RR758 pKa = 11.84RR759 pKa = 11.84QAVFPPGTDD768 pKa = 3.28LSKK771 pKa = 10.65GTSKK775 pKa = 11.04LLDD778 pKa = 3.68LAQWVPFEE786 pKa = 5.12DD787 pKa = 4.24PMTIDD792 pKa = 3.27EE793 pKa = 4.82SKK795 pKa = 10.96GLSHH799 pKa = 6.55RR800 pKa = 11.84WKK802 pKa = 10.84YY803 pKa = 10.52IFEE806 pKa = 4.46NKK808 pKa = 9.54PKK810 pKa = 10.38FLSCYY815 pKa = 8.45WDD817 pKa = 3.59EE818 pKa = 4.44NNNPIRR824 pKa = 11.84PAYY827 pKa = 9.63INLEE831 pKa = 3.91KK832 pKa = 10.85LLWPEE837 pKa = 5.22GIHH840 pKa = 5.58EE841 pKa = 4.5TIEE844 pKa = 4.41DD845 pKa = 3.93YY846 pKa = 10.87EE847 pKa = 4.18AALIGMVVDD856 pKa = 4.66NPFNHH861 pKa = 6.71HH862 pKa = 5.73NVNHH866 pKa = 6.29LMHH869 pKa = 7.4RR870 pKa = 11.84YY871 pKa = 10.01CIVQQVKK878 pKa = 10.09RR879 pKa = 11.84MMITGIKK886 pKa = 10.32AEE888 pKa = 4.58DD889 pKa = 3.45VLDD892 pKa = 4.15FCRR895 pKa = 11.84FKK897 pKa = 10.99DD898 pKa = 3.82DD899 pKa = 3.82KK900 pKa = 11.74GEE902 pKa = 4.15GVPFPMVAQWRR913 pKa = 11.84RR914 pKa = 11.84VNGWVDD920 pKa = 3.39MEE922 pKa = 4.62KK923 pKa = 10.88LPFVDD928 pKa = 5.42RR929 pKa = 11.84YY930 pKa = 9.53IAQFRR935 pKa = 11.84DD936 pKa = 3.8FVTGVTSLYY945 pKa = 10.58SRR947 pKa = 11.84SPTGGLDD954 pKa = 2.6SWRR957 pKa = 11.84FMDD960 pKa = 5.26IIRR963 pKa = 11.84GVGNLNDD970 pKa = 3.69AQFGNDD976 pKa = 2.84MVDD979 pKa = 2.16WVTFLKK985 pKa = 10.56GHH987 pKa = 7.31PLTKK991 pKa = 10.16YY992 pKa = 10.1LKK994 pKa = 7.9PTKK997 pKa = 10.16GSRR1000 pKa = 11.84KK1001 pKa = 9.32RR1002 pKa = 11.84KK1003 pKa = 8.52LHH1005 pKa = 6.36SDD1007 pKa = 3.64PEE1009 pKa = 4.36EE1010 pKa = 3.9EE1011 pKa = 3.76ALTRR1015 pKa = 11.84FKK1017 pKa = 11.2VLNHH1021 pKa = 6.07YY1022 pKa = 10.52LDD1024 pKa = 4.08PTWKK1028 pKa = 10.42KK1029 pKa = 11.38KK1030 pKa = 10.17MFDD1033 pKa = 4.05DD1034 pKa = 3.81VDD1036 pKa = 5.22SYY1038 pKa = 12.15ALWISDD1044 pKa = 3.69LLRR1047 pKa = 11.84NKK1049 pKa = 10.47GPTSS1053 pKa = 3.34
Molecular weight: 119.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1437 |
384 |
1053 |
718.5 |
80.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.299 ± 2.001 | 1.253 ± 0.093 |
6.193 ± 0.09 | 6.75 ± 0.468 |
4.245 ± 0.608 | 6.611 ± 0.07 |
2.366 ± 0.239 | 4.662 ± 0.104 |
5.776 ± 0.823 | 8.49 ± 0.757 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.714 ± 0.163 | 3.688 ± 0.096 |
5.15 ± 0.255 | 3.479 ± 0.876 |
7.794 ± 0.696 | 5.289 ± 0.609 |
5.985 ± 0.117 | 5.289 ± 0.265 |
1.949 ± 0.399 | 2.018 ± 0.43 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
