
Bambusicola thoracicus (Chinese bamboo-partridge) (Perdix thoracica)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Sarcopterygii; Dipnotetrapodomorpha; Tetrapoda; Amniota; Sauropsida; Sauria; Archelosauria; Archosauria;
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
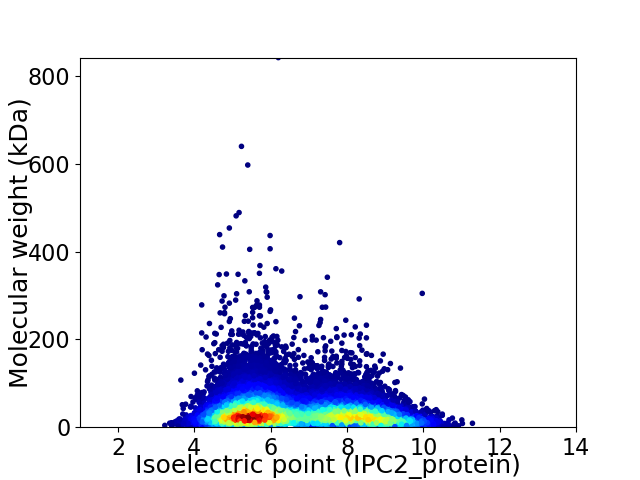
Virtual 2D-PAGE plot for 17669 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P4SU70|A0A2P4SU70_BAMTH Uncharacterized protein OS=Bambusicola thoracicus OX=9083 GN=CIB84_008584 PE=4 SV=1
NN1 pKa = 7.6CPLLPNSGQEE11 pKa = 5.08DD12 pKa = 3.93FDD14 pKa = 5.41KK15 pKa = 11.13DD16 pKa = 3.96GKK18 pKa = 11.32GDD20 pKa = 3.67ACDD23 pKa = 4.44EE24 pKa = 4.89DD25 pKa = 5.03DD26 pKa = 6.11DD27 pKa = 5.28NDD29 pKa = 4.37GVEE32 pKa = 4.65DD33 pKa = 5.47DD34 pKa = 5.49KK35 pKa = 12.08DD36 pKa = 3.71NCPLLFNPRR45 pKa = 11.84QFDD48 pKa = 3.57YY49 pKa = 11.45DD50 pKa = 3.52KK51 pKa = 11.69DD52 pKa = 4.14EE53 pKa = 4.7VGDD56 pKa = 3.79RR57 pKa = 11.84CDD59 pKa = 3.2NCPYY63 pKa = 10.11VHH65 pKa = 6.82NPAQIDD71 pKa = 3.5TDD73 pKa = 3.98NNGEE77 pKa = 3.99GDD79 pKa = 3.81SCAVDD84 pKa = 3.25IDD86 pKa = 4.11GDD88 pKa = 4.29DD89 pKa = 3.67IFNEE93 pKa = 4.17RR94 pKa = 11.84DD95 pKa = 3.22NCPYY99 pKa = 10.71VYY101 pKa = 9.85NTDD104 pKa = 3.57QSDD107 pKa = 3.51TDD109 pKa = 3.82GDD111 pKa = 4.48GVGDD115 pKa = 3.71QCDD118 pKa = 4.06NCPLMHH124 pKa = 7.14NPDD127 pKa = 3.26QTDD130 pKa = 3.12ADD132 pKa = 3.89NDD134 pKa = 4.23LVGDD138 pKa = 3.7QCDD141 pKa = 3.64NNEE144 pKa = 5.1DD145 pKa = 3.49IDD147 pKa = 4.85EE148 pKa = 5.38DD149 pKa = 3.95GHH151 pKa = 6.99QNNQDD156 pKa = 3.29NCPYY160 pKa = 10.32IPNANQADD168 pKa = 3.79HH169 pKa = 7.42DD170 pKa = 4.64KK171 pKa = 11.17DD172 pKa = 4.07GKK174 pKa = 11.27GDD176 pKa = 3.62ACDD179 pKa = 4.6PDD181 pKa = 5.2DD182 pKa = 6.11DD183 pKa = 5.43NDD185 pKa = 5.38GIPDD189 pKa = 3.84DD190 pKa = 4.96RR191 pKa = 11.84DD192 pKa = 3.43NCRR195 pKa = 11.84LRR197 pKa = 11.84YY198 pKa = 9.88NPEE201 pKa = 3.86QEE203 pKa = 5.42DD204 pKa = 3.5SDD206 pKa = 4.25GDD208 pKa = 3.89GRR210 pKa = 11.84GDD212 pKa = 3.12ICKK215 pKa = 10.38DD216 pKa = 3.51DD217 pKa = 4.78FDD219 pKa = 6.91DD220 pKa = 6.6DD221 pKa = 4.41SVPDD225 pKa = 3.82IFDD228 pKa = 3.35VCPEE232 pKa = 3.87NNAISEE238 pKa = 4.09TDD240 pKa = 3.55FRR242 pKa = 11.84KK243 pKa = 9.59FQMVPLDD250 pKa = 3.85PKK252 pKa = 10.2GTAQIDD258 pKa = 3.99PNWVIRR264 pKa = 11.84HH265 pKa = 4.99QGKK268 pKa = 9.11EE269 pKa = 3.87LVQTANSDD277 pKa = 3.25PGIAVGYY284 pKa = 10.41DD285 pKa = 3.19EE286 pKa = 5.41FSSVDD291 pKa = 3.71FSGTFYY297 pKa = 11.53VNTDD301 pKa = 3.3RR302 pKa = 11.84DD303 pKa = 3.36DD304 pKa = 4.91DD305 pKa = 4.04YY306 pKa = 12.01AGFVFGYY313 pKa = 9.43QSSSRR318 pKa = 11.84FYY320 pKa = 10.37VLMWKK325 pKa = 9.65QVTQTYY331 pKa = 8.8WEE333 pKa = 4.7DD334 pKa = 3.53KK335 pKa = 7.69PTRR338 pKa = 11.84AYY340 pKa = 10.31GYY342 pKa = 9.8SGVSLKK348 pKa = 10.51VVNSTTGTGEE358 pKa = 4.03HH359 pKa = 6.41LRR361 pKa = 11.84NALWHH366 pKa = 6.19TGNTPGQVRR375 pKa = 11.84DD376 pKa = 4.29LKK378 pKa = 10.93LHH380 pKa = 5.95HH381 pKa = 7.02CGG383 pKa = 3.92
NN1 pKa = 7.6CPLLPNSGQEE11 pKa = 5.08DD12 pKa = 3.93FDD14 pKa = 5.41KK15 pKa = 11.13DD16 pKa = 3.96GKK18 pKa = 11.32GDD20 pKa = 3.67ACDD23 pKa = 4.44EE24 pKa = 4.89DD25 pKa = 5.03DD26 pKa = 6.11DD27 pKa = 5.28NDD29 pKa = 4.37GVEE32 pKa = 4.65DD33 pKa = 5.47DD34 pKa = 5.49KK35 pKa = 12.08DD36 pKa = 3.71NCPLLFNPRR45 pKa = 11.84QFDD48 pKa = 3.57YY49 pKa = 11.45DD50 pKa = 3.52KK51 pKa = 11.69DD52 pKa = 4.14EE53 pKa = 4.7VGDD56 pKa = 3.79RR57 pKa = 11.84CDD59 pKa = 3.2NCPYY63 pKa = 10.11VHH65 pKa = 6.82NPAQIDD71 pKa = 3.5TDD73 pKa = 3.98NNGEE77 pKa = 3.99GDD79 pKa = 3.81SCAVDD84 pKa = 3.25IDD86 pKa = 4.11GDD88 pKa = 4.29DD89 pKa = 3.67IFNEE93 pKa = 4.17RR94 pKa = 11.84DD95 pKa = 3.22NCPYY99 pKa = 10.71VYY101 pKa = 9.85NTDD104 pKa = 3.57QSDD107 pKa = 3.51TDD109 pKa = 3.82GDD111 pKa = 4.48GVGDD115 pKa = 3.71QCDD118 pKa = 4.06NCPLMHH124 pKa = 7.14NPDD127 pKa = 3.26QTDD130 pKa = 3.12ADD132 pKa = 3.89NDD134 pKa = 4.23LVGDD138 pKa = 3.7QCDD141 pKa = 3.64NNEE144 pKa = 5.1DD145 pKa = 3.49IDD147 pKa = 4.85EE148 pKa = 5.38DD149 pKa = 3.95GHH151 pKa = 6.99QNNQDD156 pKa = 3.29NCPYY160 pKa = 10.32IPNANQADD168 pKa = 3.79HH169 pKa = 7.42DD170 pKa = 4.64KK171 pKa = 11.17DD172 pKa = 4.07GKK174 pKa = 11.27GDD176 pKa = 3.62ACDD179 pKa = 4.6PDD181 pKa = 5.2DD182 pKa = 6.11DD183 pKa = 5.43NDD185 pKa = 5.38GIPDD189 pKa = 3.84DD190 pKa = 4.96RR191 pKa = 11.84DD192 pKa = 3.43NCRR195 pKa = 11.84LRR197 pKa = 11.84YY198 pKa = 9.88NPEE201 pKa = 3.86QEE203 pKa = 5.42DD204 pKa = 3.5SDD206 pKa = 4.25GDD208 pKa = 3.89GRR210 pKa = 11.84GDD212 pKa = 3.12ICKK215 pKa = 10.38DD216 pKa = 3.51DD217 pKa = 4.78FDD219 pKa = 6.91DD220 pKa = 6.6DD221 pKa = 4.41SVPDD225 pKa = 3.82IFDD228 pKa = 3.35VCPEE232 pKa = 3.87NNAISEE238 pKa = 4.09TDD240 pKa = 3.55FRR242 pKa = 11.84KK243 pKa = 9.59FQMVPLDD250 pKa = 3.85PKK252 pKa = 10.2GTAQIDD258 pKa = 3.99PNWVIRR264 pKa = 11.84HH265 pKa = 4.99QGKK268 pKa = 9.11EE269 pKa = 3.87LVQTANSDD277 pKa = 3.25PGIAVGYY284 pKa = 10.41DD285 pKa = 3.19EE286 pKa = 5.41FSSVDD291 pKa = 3.71FSGTFYY297 pKa = 11.53VNTDD301 pKa = 3.3RR302 pKa = 11.84DD303 pKa = 3.36DD304 pKa = 4.91DD305 pKa = 4.04YY306 pKa = 12.01AGFVFGYY313 pKa = 9.43QSSSRR318 pKa = 11.84FYY320 pKa = 10.37VLMWKK325 pKa = 9.65QVTQTYY331 pKa = 8.8WEE333 pKa = 4.7DD334 pKa = 3.53KK335 pKa = 7.69PTRR338 pKa = 11.84AYY340 pKa = 10.31GYY342 pKa = 9.8SGVSLKK348 pKa = 10.51VVNSTTGTGEE358 pKa = 4.03HH359 pKa = 6.41LRR361 pKa = 11.84NALWHH366 pKa = 6.19TGNTPGQVRR375 pKa = 11.84DD376 pKa = 4.29LKK378 pKa = 10.93LHH380 pKa = 5.95HH381 pKa = 7.02CGG383 pKa = 3.92
Molecular weight: 42.74 kDa
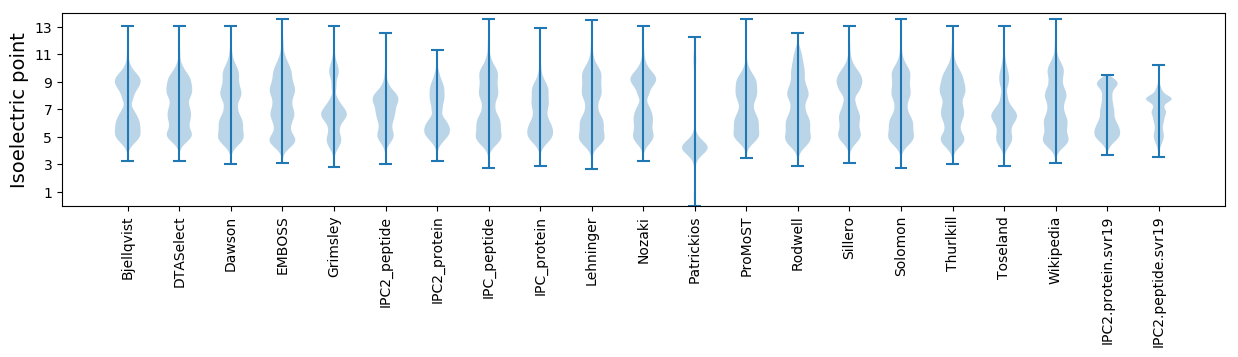
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P4SFI2|A0A2P4SFI2_BAMTH Uncharacterized protein OS=Bambusicola thoracicus OX=9083 GN=CIB84_013417 PE=3 SV=1
GG1 pKa = 7.03ARR3 pKa = 11.84RR4 pKa = 11.84SPRR7 pKa = 11.84AAAPAARR14 pKa = 11.84RR15 pKa = 11.84SAAPRR20 pKa = 11.84RR21 pKa = 11.84AAQPGSAAQRR31 pKa = 11.84PRR33 pKa = 11.84EE34 pKa = 3.96RR35 pKa = 11.84PRR37 pKa = 11.84GGRR40 pKa = 11.84GRR42 pKa = 11.84RR43 pKa = 11.84ARR45 pKa = 11.84VPPRR49 pKa = 11.84QLRR52 pKa = 11.84RR53 pKa = 11.84GLRR56 pKa = 11.84GVLLVRR62 pKa = 11.84RR63 pKa = 11.84APHH66 pKa = 5.59QGGVPPLRR74 pKa = 11.84HH75 pKa = 6.12PGAARR80 pKa = 11.84RR81 pKa = 11.84RR82 pKa = 3.88
GG1 pKa = 7.03ARR3 pKa = 11.84RR4 pKa = 11.84SPRR7 pKa = 11.84AAAPAARR14 pKa = 11.84RR15 pKa = 11.84SAAPRR20 pKa = 11.84RR21 pKa = 11.84AAQPGSAAQRR31 pKa = 11.84PRR33 pKa = 11.84EE34 pKa = 3.96RR35 pKa = 11.84PRR37 pKa = 11.84GGRR40 pKa = 11.84GRR42 pKa = 11.84RR43 pKa = 11.84ARR45 pKa = 11.84VPPRR49 pKa = 11.84QLRR52 pKa = 11.84RR53 pKa = 11.84GLRR56 pKa = 11.84GVLLVRR62 pKa = 11.84RR63 pKa = 11.84APHH66 pKa = 5.59QGGVPPLRR74 pKa = 11.84HH75 pKa = 6.12PGAARR80 pKa = 11.84RR81 pKa = 11.84RR82 pKa = 3.88
Molecular weight: 8.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6035687 |
8 |
7645 |
341.6 |
38.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.096 ± 0.024 | 2.319 ± 0.015 |
4.867 ± 0.013 | 7.171 ± 0.029 |
3.766 ± 0.016 | 6.23 ± 0.025 |
2.519 ± 0.01 | 4.579 ± 0.016 |
5.928 ± 0.025 | 9.772 ± 0.027 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.25 ± 0.008 | 3.819 ± 0.018 |
5.658 ± 0.025 | 4.605 ± 0.019 |
5.457 ± 0.018 | 8.322 ± 0.027 |
5.281 ± 0.016 | 6.32 ± 0.018 |
1.187 ± 0.008 | 2.843 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
