
Aphanothece sacrum FPU1
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Oscillatoriophycideae; Chroococcales; Aphanothecaceae; Aphanothece; Aphanothece sacrum
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
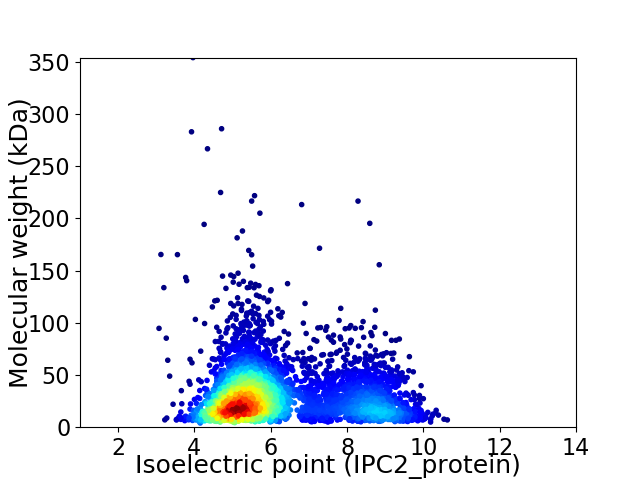
Virtual 2D-PAGE plot for 4301 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A401IC74|A0A401IC74_APHSA Ferredoxin-nitrite reductase OS=Aphanothece sacrum FPU1 OX=1920663 GN=AsFPU1_0239 PE=4 SV=1
MM1 pKa = 7.53AVLEE5 pKa = 4.19QAFLLSQTTLQNFANNPDD23 pKa = 3.83FAAKK27 pKa = 8.85MALAFNSTSQADD39 pKa = 4.02TLQTAWLNGDD49 pKa = 3.59FAGFPALEE57 pKa = 3.68IRR59 pKa = 11.84YY60 pKa = 9.29QSEE63 pKa = 4.21LNGANGAYY71 pKa = 10.17SADD74 pKa = 3.2TGIIYY79 pKa = 10.38LAYY82 pKa = 9.87EE83 pKa = 4.18YY84 pKa = 10.71LLQLFPPSQGGLGGINALVGLILEE108 pKa = 4.7EE109 pKa = 4.13YY110 pKa = 8.63GHH112 pKa = 6.07YY113 pKa = 10.62VDD115 pKa = 5.83VVLNGSNDD123 pKa = 3.36SDD125 pKa = 3.88GDD127 pKa = 3.47EE128 pKa = 4.21GAIFSALVMGQSLSEE143 pKa = 4.07TDD145 pKa = 3.44LGQLKK150 pKa = 9.08TEE152 pKa = 4.3NDD154 pKa = 3.23HH155 pKa = 7.1GVISFNGVNIAVEE168 pKa = 4.22MQNFAGSAGNDD179 pKa = 3.9TITGTSGDD187 pKa = 4.16DD188 pKa = 3.78VIDD191 pKa = 4.02GLGGNDD197 pKa = 4.15SLNGLAGDD205 pKa = 5.25DD206 pKa = 4.02ILNGGDD212 pKa = 4.55GNDD215 pKa = 3.94TLDD218 pKa = 4.22GGDD221 pKa = 4.54GNDD224 pKa = 3.63ILNPGLGVDD233 pKa = 4.5TIDD236 pKa = 4.52GNTGTDD242 pKa = 3.51TLIADD247 pKa = 3.89YY248 pKa = 7.79TTATVSGIGTNNYY261 pKa = 10.21GITNTSSKK269 pKa = 10.07IYY271 pKa = 10.22SSYY274 pKa = 11.61SSTTLLNYY282 pKa = 10.81SNIEE286 pKa = 3.94LFNITGTQYY295 pKa = 11.54NDD297 pKa = 3.08SFSANTGYY305 pKa = 10.68KK306 pKa = 9.5INAGGGSDD314 pKa = 3.77TLTLSLSNTSTNWVLDD330 pKa = 3.51WANANAQLSGDD341 pKa = 4.06SNTQISNFEE350 pKa = 4.61KK351 pKa = 10.26ISSVTTGSGNDD362 pKa = 3.2TFKK365 pKa = 10.53FTNYY369 pKa = 10.16LLNSSIDD376 pKa = 3.72GNTGTDD382 pKa = 3.48TLIADD387 pKa = 4.29CTTATVSGIGTNNYY401 pKa = 10.21GITNTSSKK409 pKa = 10.07IYY411 pKa = 10.22SSYY414 pKa = 11.61SSTTLLNYY422 pKa = 10.81SNIEE426 pKa = 3.94LFNITGTQYY435 pKa = 11.23NDD437 pKa = 3.16SLTGYY442 pKa = 9.91AGNDD446 pKa = 3.64TLIGAAGDD454 pKa = 3.63DD455 pKa = 4.55TINGTIGDD463 pKa = 4.35SLDD466 pKa = 3.71GGEE469 pKa = 4.55GTDD472 pKa = 3.87TVNLDD477 pKa = 3.78LSSAMTNLTFDD488 pKa = 4.68LDD490 pKa = 4.24LSVTTNLQGISGTEE504 pKa = 3.5VKK506 pKa = 10.61NFEE509 pKa = 4.63KK510 pKa = 10.34IGTITTGSGNDD521 pKa = 3.2TFKK524 pKa = 10.5FTNYY528 pKa = 10.65LLGGTINGNTGTDD541 pKa = 3.56TLIADD546 pKa = 3.92YY547 pKa = 7.79TTATGSGISTNNYY560 pKa = 9.62GITNTSSRR568 pKa = 11.84IYY570 pKa = 10.53SSYY573 pKa = 11.49NSGTFLNYY581 pKa = 10.77SNIEE585 pKa = 4.02LFNITGTQYY594 pKa = 11.23NDD596 pKa = 3.16SLTGYY601 pKa = 9.91AGNDD605 pKa = 3.64TLIGAAGDD613 pKa = 3.63DD614 pKa = 4.55TINGTIGDD622 pKa = 4.43SLDD625 pKa = 3.63GGKK628 pKa = 8.37GTDD631 pKa = 3.56TVNLDD636 pKa = 3.53LSSAITNLTFDD647 pKa = 4.96LDD649 pKa = 4.24LSVTTNLQGISGTEE663 pKa = 3.5VKK665 pKa = 10.61NFEE668 pKa = 4.63KK669 pKa = 10.34IGTITTGSGNDD680 pKa = 3.2TFKK683 pKa = 10.5FTNYY687 pKa = 10.65LLGGTINGNTGTDD700 pKa = 3.56TLIADD705 pKa = 3.92YY706 pKa = 7.79TTATGSGISTNNYY719 pKa = 9.62GITNTSSRR727 pKa = 11.84IYY729 pKa = 10.53SSYY732 pKa = 11.49NSGTFLNYY740 pKa = 10.77SNIEE744 pKa = 4.02LFNITGTQYY753 pKa = 11.23NDD755 pKa = 3.16SLTGYY760 pKa = 9.91AGNDD764 pKa = 3.64TLIGAAGDD772 pKa = 3.63DD773 pKa = 4.55TINGTIGDD781 pKa = 4.43SLDD784 pKa = 3.63GGKK787 pKa = 8.37GTDD790 pKa = 3.56TVNLDD795 pKa = 3.53LSSAITNLTFDD806 pKa = 4.96LDD808 pKa = 4.24LSVTTNLQGISGTEE822 pKa = 3.5VKK824 pKa = 10.61NFEE827 pKa = 4.63KK828 pKa = 10.34IGTITTGSGNDD839 pKa = 3.2TFKK842 pKa = 10.5FTNYY846 pKa = 10.65LLGGTINGNTGTDD859 pKa = 3.56TLIADD864 pKa = 3.92YY865 pKa = 7.79TTATGSGISTNNYY878 pKa = 9.62GITNTSSKK886 pKa = 10.07IYY888 pKa = 10.22SSYY891 pKa = 11.61SSTTLLNYY899 pKa = 10.81SNIEE903 pKa = 3.94LFNITGTQYY912 pKa = 11.53NDD914 pKa = 3.22NLNGSGGNDD923 pKa = 3.3TLIGGEE929 pKa = 4.47GNDD932 pKa = 4.04SLSGGDD938 pKa = 4.31GNDD941 pKa = 3.26KK942 pKa = 11.01LSGVNAKK949 pKa = 10.35KK950 pKa = 9.09LTPGLNEE957 pKa = 4.03IDD959 pKa = 3.8TLSGGNSIDD968 pKa = 3.63TFILGDD974 pKa = 3.67AANIYY979 pKa = 10.81YY980 pKa = 10.08DD981 pKa = 4.16DD982 pKa = 4.79RR983 pKa = 11.84NNTTTGTTDD992 pKa = 3.17YY993 pKa = 11.78ARR995 pKa = 11.84ITDD998 pKa = 4.48FNATQDD1004 pKa = 3.71IIQLNGAKK1012 pKa = 10.02SNYY1015 pKa = 9.71RR1016 pKa = 11.84LTTASFNNISGTAIYY1031 pKa = 10.35VDD1033 pKa = 4.14KK1034 pKa = 10.97PNTEE1038 pKa = 3.95PDD1040 pKa = 3.33EE1041 pKa = 4.77LIGFIEE1047 pKa = 4.93GVTGLDD1053 pKa = 3.05INSNAFVTAKK1063 pKa = 10.97DD1064 pKa = 3.7EE1065 pKa = 4.34IAFSNAQFRR1074 pKa = 11.84VIEE1077 pKa = 4.8DD1078 pKa = 3.57GTPVVAVTVVRR1089 pKa = 11.84SGAVQTEE1096 pKa = 4.06VSATITLNNGTAISPEE1112 pKa = 4.47DD1113 pKa = 3.9YY1114 pKa = 10.63NNTPIIVNFAQGEE1127 pKa = 4.28TSQTVIIPIIDD1138 pKa = 3.8DD1139 pKa = 3.99SEE1141 pKa = 4.7FEE1143 pKa = 4.18LDD1145 pKa = 3.29EE1146 pKa = 4.86TINLTLTNPTNGAVLGSQNTASLTIVDD1173 pKa = 4.59NEE1175 pKa = 4.09IPLPGILAFSATNYY1189 pKa = 10.58SITEE1193 pKa = 4.41DD1194 pKa = 3.38GTPIVTVTIIRR1205 pKa = 11.84TDD1207 pKa = 3.41GSDD1210 pKa = 3.46GAVSATLNLSDD1221 pKa = 5.04DD1222 pKa = 3.93TATAPNDD1229 pKa = 3.59YY1230 pKa = 10.33GNTPIVVNFAHH1241 pKa = 7.46RR1242 pKa = 11.84EE1243 pKa = 3.79TSKK1246 pKa = 9.93TVTIPIVNDD1255 pKa = 3.82SIYY1258 pKa = 11.04EE1259 pKa = 4.0GDD1261 pKa = 3.65EE1262 pKa = 4.05TLKK1265 pKa = 11.15LSLTNPTGGATLGGQNTANLTIVDD1289 pKa = 4.14NDD1291 pKa = 3.94LPTISLVVSPDD1302 pKa = 3.1SVTEE1306 pKa = 4.35DD1307 pKa = 3.13GLTNLVYY1314 pKa = 10.42TFIRR1318 pKa = 11.84TGNTVSPLTVNFNVGGTGIFNNDD1341 pKa = 3.79YY1342 pKa = 9.27IQSGAASFNGTSGSITFAAGSDD1364 pKa = 3.76TAILTLDD1371 pKa = 3.56PTVDD1375 pKa = 3.33SMFEE1379 pKa = 3.69ADD1381 pKa = 3.42EE1382 pKa = 4.48TVNLTLVSSANYY1394 pKa = 10.02NRR1396 pKa = 11.84GTTTAVNSIITNDD1409 pKa = 4.65DD1410 pKa = 3.32INSGTFSFSSPQFTVNEE1427 pKa = 4.57DD1428 pKa = 3.49GTPITAVTINRR1439 pKa = 11.84AGNSNGEE1446 pKa = 4.02VSVTINLNNGTATAPNDD1463 pKa = 3.76YY1464 pKa = 9.79NNSPIVVTFASGQQTKK1480 pKa = 7.92TVNIPIQNDD1489 pKa = 3.3LTRR1492 pKa = 11.84EE1493 pKa = 3.89GDD1495 pKa = 3.29EE1496 pKa = 4.63SINMVLSNPTGGANLGTQTSANLIIADD1523 pKa = 3.91NDD1525 pKa = 3.13IGLNAEE1531 pKa = 4.12YY1532 pKa = 10.65FNGYY1536 pKa = 10.23FNDD1539 pKa = 3.41NLGFFTANQPILKK1552 pKa = 8.95RR1553 pKa = 11.84TDD1555 pKa = 2.93KK1556 pKa = 10.67TVNFLNLAWGHH1567 pKa = 5.35EE1568 pKa = 4.33TLKK1571 pKa = 11.02ALPDD1575 pKa = 3.91DD1576 pKa = 4.77EE1577 pKa = 6.11KK1578 pKa = 11.67SKK1580 pKa = 8.9TWW1582 pKa = 3.13
MM1 pKa = 7.53AVLEE5 pKa = 4.19QAFLLSQTTLQNFANNPDD23 pKa = 3.83FAAKK27 pKa = 8.85MALAFNSTSQADD39 pKa = 4.02TLQTAWLNGDD49 pKa = 3.59FAGFPALEE57 pKa = 3.68IRR59 pKa = 11.84YY60 pKa = 9.29QSEE63 pKa = 4.21LNGANGAYY71 pKa = 10.17SADD74 pKa = 3.2TGIIYY79 pKa = 10.38LAYY82 pKa = 9.87EE83 pKa = 4.18YY84 pKa = 10.71LLQLFPPSQGGLGGINALVGLILEE108 pKa = 4.7EE109 pKa = 4.13YY110 pKa = 8.63GHH112 pKa = 6.07YY113 pKa = 10.62VDD115 pKa = 5.83VVLNGSNDD123 pKa = 3.36SDD125 pKa = 3.88GDD127 pKa = 3.47EE128 pKa = 4.21GAIFSALVMGQSLSEE143 pKa = 4.07TDD145 pKa = 3.44LGQLKK150 pKa = 9.08TEE152 pKa = 4.3NDD154 pKa = 3.23HH155 pKa = 7.1GVISFNGVNIAVEE168 pKa = 4.22MQNFAGSAGNDD179 pKa = 3.9TITGTSGDD187 pKa = 4.16DD188 pKa = 3.78VIDD191 pKa = 4.02GLGGNDD197 pKa = 4.15SLNGLAGDD205 pKa = 5.25DD206 pKa = 4.02ILNGGDD212 pKa = 4.55GNDD215 pKa = 3.94TLDD218 pKa = 4.22GGDD221 pKa = 4.54GNDD224 pKa = 3.63ILNPGLGVDD233 pKa = 4.5TIDD236 pKa = 4.52GNTGTDD242 pKa = 3.51TLIADD247 pKa = 3.89YY248 pKa = 7.79TTATVSGIGTNNYY261 pKa = 10.21GITNTSSKK269 pKa = 10.07IYY271 pKa = 10.22SSYY274 pKa = 11.61SSTTLLNYY282 pKa = 10.81SNIEE286 pKa = 3.94LFNITGTQYY295 pKa = 11.54NDD297 pKa = 3.08SFSANTGYY305 pKa = 10.68KK306 pKa = 9.5INAGGGSDD314 pKa = 3.77TLTLSLSNTSTNWVLDD330 pKa = 3.51WANANAQLSGDD341 pKa = 4.06SNTQISNFEE350 pKa = 4.61KK351 pKa = 10.26ISSVTTGSGNDD362 pKa = 3.2TFKK365 pKa = 10.53FTNYY369 pKa = 10.16LLNSSIDD376 pKa = 3.72GNTGTDD382 pKa = 3.48TLIADD387 pKa = 4.29CTTATVSGIGTNNYY401 pKa = 10.21GITNTSSKK409 pKa = 10.07IYY411 pKa = 10.22SSYY414 pKa = 11.61SSTTLLNYY422 pKa = 10.81SNIEE426 pKa = 3.94LFNITGTQYY435 pKa = 11.23NDD437 pKa = 3.16SLTGYY442 pKa = 9.91AGNDD446 pKa = 3.64TLIGAAGDD454 pKa = 3.63DD455 pKa = 4.55TINGTIGDD463 pKa = 4.35SLDD466 pKa = 3.71GGEE469 pKa = 4.55GTDD472 pKa = 3.87TVNLDD477 pKa = 3.78LSSAMTNLTFDD488 pKa = 4.68LDD490 pKa = 4.24LSVTTNLQGISGTEE504 pKa = 3.5VKK506 pKa = 10.61NFEE509 pKa = 4.63KK510 pKa = 10.34IGTITTGSGNDD521 pKa = 3.2TFKK524 pKa = 10.5FTNYY528 pKa = 10.65LLGGTINGNTGTDD541 pKa = 3.56TLIADD546 pKa = 3.92YY547 pKa = 7.79TTATGSGISTNNYY560 pKa = 9.62GITNTSSRR568 pKa = 11.84IYY570 pKa = 10.53SSYY573 pKa = 11.49NSGTFLNYY581 pKa = 10.77SNIEE585 pKa = 4.02LFNITGTQYY594 pKa = 11.23NDD596 pKa = 3.16SLTGYY601 pKa = 9.91AGNDD605 pKa = 3.64TLIGAAGDD613 pKa = 3.63DD614 pKa = 4.55TINGTIGDD622 pKa = 4.43SLDD625 pKa = 3.63GGKK628 pKa = 8.37GTDD631 pKa = 3.56TVNLDD636 pKa = 3.53LSSAITNLTFDD647 pKa = 4.96LDD649 pKa = 4.24LSVTTNLQGISGTEE663 pKa = 3.5VKK665 pKa = 10.61NFEE668 pKa = 4.63KK669 pKa = 10.34IGTITTGSGNDD680 pKa = 3.2TFKK683 pKa = 10.5FTNYY687 pKa = 10.65LLGGTINGNTGTDD700 pKa = 3.56TLIADD705 pKa = 3.92YY706 pKa = 7.79TTATGSGISTNNYY719 pKa = 9.62GITNTSSRR727 pKa = 11.84IYY729 pKa = 10.53SSYY732 pKa = 11.49NSGTFLNYY740 pKa = 10.77SNIEE744 pKa = 4.02LFNITGTQYY753 pKa = 11.23NDD755 pKa = 3.16SLTGYY760 pKa = 9.91AGNDD764 pKa = 3.64TLIGAAGDD772 pKa = 3.63DD773 pKa = 4.55TINGTIGDD781 pKa = 4.43SLDD784 pKa = 3.63GGKK787 pKa = 8.37GTDD790 pKa = 3.56TVNLDD795 pKa = 3.53LSSAITNLTFDD806 pKa = 4.96LDD808 pKa = 4.24LSVTTNLQGISGTEE822 pKa = 3.5VKK824 pKa = 10.61NFEE827 pKa = 4.63KK828 pKa = 10.34IGTITTGSGNDD839 pKa = 3.2TFKK842 pKa = 10.5FTNYY846 pKa = 10.65LLGGTINGNTGTDD859 pKa = 3.56TLIADD864 pKa = 3.92YY865 pKa = 7.79TTATGSGISTNNYY878 pKa = 9.62GITNTSSKK886 pKa = 10.07IYY888 pKa = 10.22SSYY891 pKa = 11.61SSTTLLNYY899 pKa = 10.81SNIEE903 pKa = 3.94LFNITGTQYY912 pKa = 11.53NDD914 pKa = 3.22NLNGSGGNDD923 pKa = 3.3TLIGGEE929 pKa = 4.47GNDD932 pKa = 4.04SLSGGDD938 pKa = 4.31GNDD941 pKa = 3.26KK942 pKa = 11.01LSGVNAKK949 pKa = 10.35KK950 pKa = 9.09LTPGLNEE957 pKa = 4.03IDD959 pKa = 3.8TLSGGNSIDD968 pKa = 3.63TFILGDD974 pKa = 3.67AANIYY979 pKa = 10.81YY980 pKa = 10.08DD981 pKa = 4.16DD982 pKa = 4.79RR983 pKa = 11.84NNTTTGTTDD992 pKa = 3.17YY993 pKa = 11.78ARR995 pKa = 11.84ITDD998 pKa = 4.48FNATQDD1004 pKa = 3.71IIQLNGAKK1012 pKa = 10.02SNYY1015 pKa = 9.71RR1016 pKa = 11.84LTTASFNNISGTAIYY1031 pKa = 10.35VDD1033 pKa = 4.14KK1034 pKa = 10.97PNTEE1038 pKa = 3.95PDD1040 pKa = 3.33EE1041 pKa = 4.77LIGFIEE1047 pKa = 4.93GVTGLDD1053 pKa = 3.05INSNAFVTAKK1063 pKa = 10.97DD1064 pKa = 3.7EE1065 pKa = 4.34IAFSNAQFRR1074 pKa = 11.84VIEE1077 pKa = 4.8DD1078 pKa = 3.57GTPVVAVTVVRR1089 pKa = 11.84SGAVQTEE1096 pKa = 4.06VSATITLNNGTAISPEE1112 pKa = 4.47DD1113 pKa = 3.9YY1114 pKa = 10.63NNTPIIVNFAQGEE1127 pKa = 4.28TSQTVIIPIIDD1138 pKa = 3.8DD1139 pKa = 3.99SEE1141 pKa = 4.7FEE1143 pKa = 4.18LDD1145 pKa = 3.29EE1146 pKa = 4.86TINLTLTNPTNGAVLGSQNTASLTIVDD1173 pKa = 4.59NEE1175 pKa = 4.09IPLPGILAFSATNYY1189 pKa = 10.58SITEE1193 pKa = 4.41DD1194 pKa = 3.38GTPIVTVTIIRR1205 pKa = 11.84TDD1207 pKa = 3.41GSDD1210 pKa = 3.46GAVSATLNLSDD1221 pKa = 5.04DD1222 pKa = 3.93TATAPNDD1229 pKa = 3.59YY1230 pKa = 10.33GNTPIVVNFAHH1241 pKa = 7.46RR1242 pKa = 11.84EE1243 pKa = 3.79TSKK1246 pKa = 9.93TVTIPIVNDD1255 pKa = 3.82SIYY1258 pKa = 11.04EE1259 pKa = 4.0GDD1261 pKa = 3.65EE1262 pKa = 4.05TLKK1265 pKa = 11.15LSLTNPTGGATLGGQNTANLTIVDD1289 pKa = 4.14NDD1291 pKa = 3.94LPTISLVVSPDD1302 pKa = 3.1SVTEE1306 pKa = 4.35DD1307 pKa = 3.13GLTNLVYY1314 pKa = 10.42TFIRR1318 pKa = 11.84TGNTVSPLTVNFNVGGTGIFNNDD1341 pKa = 3.79YY1342 pKa = 9.27IQSGAASFNGTSGSITFAAGSDD1364 pKa = 3.76TAILTLDD1371 pKa = 3.56PTVDD1375 pKa = 3.33SMFEE1379 pKa = 3.69ADD1381 pKa = 3.42EE1382 pKa = 4.48TVNLTLVSSANYY1394 pKa = 10.02NRR1396 pKa = 11.84GTTTAVNSIITNDD1409 pKa = 4.65DD1410 pKa = 3.32INSGTFSFSSPQFTVNEE1427 pKa = 4.57DD1428 pKa = 3.49GTPITAVTINRR1439 pKa = 11.84AGNSNGEE1446 pKa = 4.02VSVTINLNNGTATAPNDD1463 pKa = 3.76YY1464 pKa = 9.79NNSPIVVTFASGQQTKK1480 pKa = 7.92TVNIPIQNDD1489 pKa = 3.3LTRR1492 pKa = 11.84EE1493 pKa = 3.89GDD1495 pKa = 3.29EE1496 pKa = 4.63SINMVLSNPTGGANLGTQTSANLIIADD1523 pKa = 3.91NDD1525 pKa = 3.13IGLNAEE1531 pKa = 4.12YY1532 pKa = 10.65FNGYY1536 pKa = 10.23FNDD1539 pKa = 3.41NLGFFTANQPILKK1552 pKa = 8.95RR1553 pKa = 11.84TDD1555 pKa = 2.93KK1556 pKa = 10.67TVNFLNLAWGHH1567 pKa = 5.35EE1568 pKa = 4.33TLKK1571 pKa = 11.02ALPDD1575 pKa = 3.91DD1576 pKa = 4.77EE1577 pKa = 6.11KK1578 pKa = 11.67SKK1580 pKa = 8.9TWW1582 pKa = 3.13
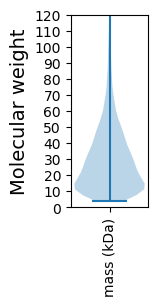
Molecular weight: 165.27 kDa
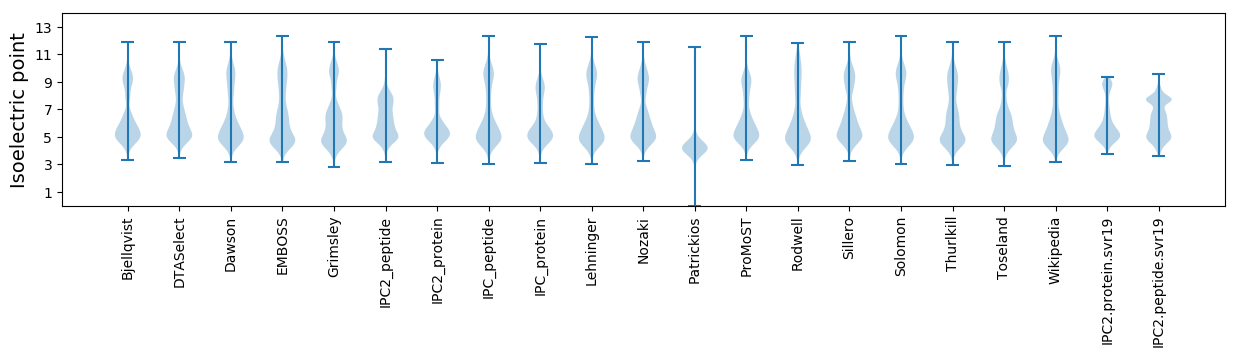
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A401IKL5|A0A401IKL5_APHSA VWFA domain-containing protein OS=Aphanothece sacrum FPU1 OX=1920663 GN=AsFPU1_3140 PE=4 SV=1
MM1 pKa = 7.78IEE3 pKa = 4.04RR4 pKa = 11.84EE5 pKa = 4.23KK6 pKa = 10.96KK7 pKa = 9.86RR8 pKa = 11.84SRR10 pKa = 11.84LVDD13 pKa = 3.59KK14 pKa = 10.8YY15 pKa = 10.75AEE17 pKa = 3.97KK18 pKa = 10.6RR19 pKa = 11.84ATLKK23 pKa = 11.02EE24 pKa = 4.07EE25 pKa = 3.78FRR27 pKa = 11.84VAEE30 pKa = 4.53DD31 pKa = 4.44YY32 pKa = 10.56EE33 pKa = 5.33DD34 pKa = 3.73KK35 pKa = 11.02MNLQRR40 pKa = 11.84QLQQLPRR47 pKa = 11.84NSSPNRR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84NRR57 pKa = 11.84CWVTGRR63 pKa = 11.84SRR65 pKa = 11.84GYY67 pKa = 10.25YY68 pKa = 9.53RR69 pKa = 11.84DD70 pKa = 3.76FGLSRR75 pKa = 11.84NVLRR79 pKa = 11.84EE80 pKa = 3.67WAHH83 pKa = 5.74QGLLPGVVKK92 pKa = 10.78SSWW95 pKa = 2.92
MM1 pKa = 7.78IEE3 pKa = 4.04RR4 pKa = 11.84EE5 pKa = 4.23KK6 pKa = 10.96KK7 pKa = 9.86RR8 pKa = 11.84SRR10 pKa = 11.84LVDD13 pKa = 3.59KK14 pKa = 10.8YY15 pKa = 10.75AEE17 pKa = 3.97KK18 pKa = 10.6RR19 pKa = 11.84ATLKK23 pKa = 11.02EE24 pKa = 4.07EE25 pKa = 3.78FRR27 pKa = 11.84VAEE30 pKa = 4.53DD31 pKa = 4.44YY32 pKa = 10.56EE33 pKa = 5.33DD34 pKa = 3.73KK35 pKa = 11.02MNLQRR40 pKa = 11.84QLQQLPRR47 pKa = 11.84NSSPNRR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84NRR57 pKa = 11.84CWVTGRR63 pKa = 11.84SRR65 pKa = 11.84GYY67 pKa = 10.25YY68 pKa = 9.53RR69 pKa = 11.84DD70 pKa = 3.76FGLSRR75 pKa = 11.84NVLRR79 pKa = 11.84EE80 pKa = 3.67WAHH83 pKa = 5.74QGLLPGVVKK92 pKa = 10.78SSWW95 pKa = 2.92
Molecular weight: 11.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1220765 |
39 |
3314 |
283.8 |
31.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.634 ± 0.041 | 0.992 ± 0.015 |
4.949 ± 0.036 | 6.53 ± 0.044 |
4.058 ± 0.029 | 6.57 ± 0.051 |
1.757 ± 0.021 | 7.763 ± 0.035 |
5.572 ± 0.042 | 11.128 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.917 ± 0.02 | 4.913 ± 0.044 |
4.563 ± 0.03 | 5.293 ± 0.036 |
4.521 ± 0.036 | 6.36 ± 0.033 |
5.757 ± 0.047 | 6.028 ± 0.032 |
1.413 ± 0.019 | 3.28 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
