
Streptococcus phage Javan536
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Moineauvirus; unclassified Moineauvirus
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
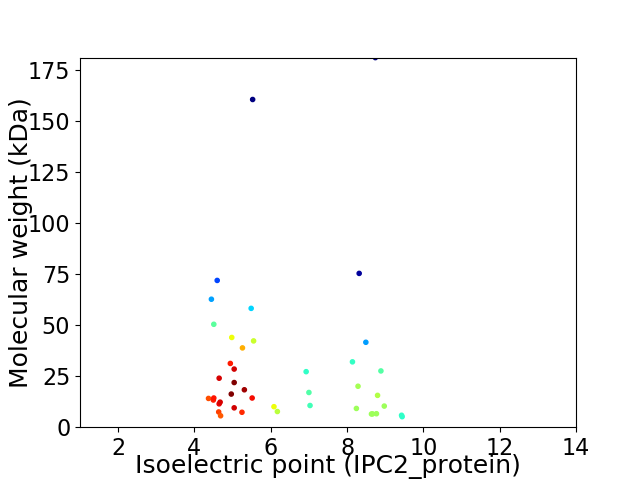
Virtual 2D-PAGE plot for 43 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D6BEU3|A0A4D6BEU3_9CAUD Uncharacterized protein OS=Streptococcus phage Javan536 OX=2548249 GN=Javan536_0008 PE=4 SV=1
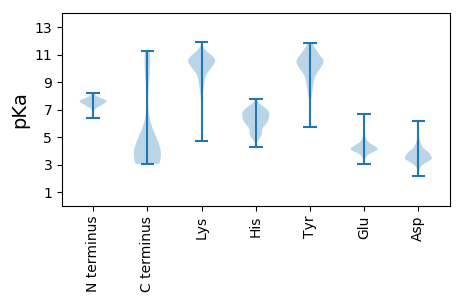
MM1 pKa = 7.17TSKK4 pKa = 8.78TQLLNTLEE12 pKa = 4.22SLVNQRR18 pKa = 11.84VTVPTNPYY26 pKa = 9.94GGQCAALIDD35 pKa = 4.29YY36 pKa = 8.32VLQYY40 pKa = 11.52AGLFNLDD47 pKa = 3.78FSYY50 pKa = 11.08MNAIDD55 pKa = 4.08GLDD58 pKa = 3.27RR59 pKa = 11.84AEE61 pKa = 4.02NLGLKK66 pKa = 7.36VTRR69 pKa = 11.84FNGSNNPPVGSVWVTNCLPYY89 pKa = 10.51HH90 pKa = 5.36QFGHH94 pKa = 6.24IGFVVAEE101 pKa = 4.33NPDD104 pKa = 3.56GTVTTVEE111 pKa = 3.96QNIDD115 pKa = 3.34GNADD119 pKa = 3.43CLYY122 pKa = 10.67NGGWTRR128 pKa = 11.84KK129 pKa = 5.87VTRR132 pKa = 11.84NLDD135 pKa = 3.39SAGNFSYY142 pKa = 10.63IDD144 pKa = 3.24WSAPSQQMVGWFEE157 pKa = 4.62LPFDD161 pKa = 4.39GMTEE165 pKa = 3.7NNYY168 pKa = 10.45FIDD171 pKa = 3.6VSAYY175 pKa = 9.87QPGDD179 pKa = 3.36LTSICSASGTNNTVIKK195 pKa = 9.37VTEE198 pKa = 4.14GVGWVSPVATQQTNTSNCIGYY219 pKa = 7.92YY220 pKa = 9.85HH221 pKa = 7.1FARR224 pKa = 11.84FGGDD228 pKa = 2.81VATAQAEE235 pKa = 4.18ANYY238 pKa = 9.8FISNLPSHH246 pKa = 6.82PRR248 pKa = 11.84YY249 pKa = 9.38LVCDD253 pKa = 4.07YY254 pKa = 11.32EE255 pKa = 6.54DD256 pKa = 4.01GASGDD261 pKa = 3.62KK262 pKa = 10.06QANTNAVLAFMDD274 pKa = 3.3ICKK277 pKa = 10.71ANGFEE282 pKa = 4.51PIYY285 pKa = 10.66YY286 pKa = 9.81SYY288 pKa = 11.36KK289 pKa = 10.34PYY291 pKa = 10.33TLANVYY297 pKa = 9.91VDD299 pKa = 4.33QITARR304 pKa = 11.84YY305 pKa = 7.8PNSLWIAAYY314 pKa = 8.94PDD316 pKa = 3.74YY317 pKa = 10.64EE318 pKa = 4.52VRR320 pKa = 11.84PEE322 pKa = 4.38PYY324 pKa = 9.07WGVYY328 pKa = 10.28PNMEE332 pKa = 4.23HH333 pKa = 5.47TRR335 pKa = 11.84WWQFTSTGLAGGLDD349 pKa = 3.76KK350 pKa = 11.39NVVIINDD357 pKa = 3.56GDD359 pKa = 4.01SLVNKK364 pKa = 9.9EE365 pKa = 4.19KK366 pKa = 11.01EE367 pKa = 4.39EE368 pKa = 4.09EE369 pKa = 4.06NMDD372 pKa = 3.59YY373 pKa = 10.92VVRR376 pKa = 11.84SEE378 pKa = 4.67SGSQGYY384 pKa = 10.02VGVVNGRR391 pKa = 11.84VFGIGSMGTVDD402 pKa = 3.97ALRR405 pKa = 11.84SAGAKK410 pKa = 10.07HH411 pKa = 5.78LTLPDD416 pKa = 3.46ADD418 pKa = 3.99FDD420 pKa = 4.53RR421 pKa = 11.84FLNSQSNDD429 pKa = 3.02TQAVAKK435 pKa = 10.01AVSEE439 pKa = 4.22ASASVVKK446 pKa = 10.68AIEE449 pKa = 3.89EE450 pKa = 4.05RR451 pKa = 11.84AQATQGQTGVV461 pKa = 3.31
MM1 pKa = 7.17TSKK4 pKa = 8.78TQLLNTLEE12 pKa = 4.22SLVNQRR18 pKa = 11.84VTVPTNPYY26 pKa = 9.94GGQCAALIDD35 pKa = 4.29YY36 pKa = 8.32VLQYY40 pKa = 11.52AGLFNLDD47 pKa = 3.78FSYY50 pKa = 11.08MNAIDD55 pKa = 4.08GLDD58 pKa = 3.27RR59 pKa = 11.84AEE61 pKa = 4.02NLGLKK66 pKa = 7.36VTRR69 pKa = 11.84FNGSNNPPVGSVWVTNCLPYY89 pKa = 10.51HH90 pKa = 5.36QFGHH94 pKa = 6.24IGFVVAEE101 pKa = 4.33NPDD104 pKa = 3.56GTVTTVEE111 pKa = 3.96QNIDD115 pKa = 3.34GNADD119 pKa = 3.43CLYY122 pKa = 10.67NGGWTRR128 pKa = 11.84KK129 pKa = 5.87VTRR132 pKa = 11.84NLDD135 pKa = 3.39SAGNFSYY142 pKa = 10.63IDD144 pKa = 3.24WSAPSQQMVGWFEE157 pKa = 4.62LPFDD161 pKa = 4.39GMTEE165 pKa = 3.7NNYY168 pKa = 10.45FIDD171 pKa = 3.6VSAYY175 pKa = 9.87QPGDD179 pKa = 3.36LTSICSASGTNNTVIKK195 pKa = 9.37VTEE198 pKa = 4.14GVGWVSPVATQQTNTSNCIGYY219 pKa = 7.92YY220 pKa = 9.85HH221 pKa = 7.1FARR224 pKa = 11.84FGGDD228 pKa = 2.81VATAQAEE235 pKa = 4.18ANYY238 pKa = 9.8FISNLPSHH246 pKa = 6.82PRR248 pKa = 11.84YY249 pKa = 9.38LVCDD253 pKa = 4.07YY254 pKa = 11.32EE255 pKa = 6.54DD256 pKa = 4.01GASGDD261 pKa = 3.62KK262 pKa = 10.06QANTNAVLAFMDD274 pKa = 3.3ICKK277 pKa = 10.71ANGFEE282 pKa = 4.51PIYY285 pKa = 10.66YY286 pKa = 9.81SYY288 pKa = 11.36KK289 pKa = 10.34PYY291 pKa = 10.33TLANVYY297 pKa = 9.91VDD299 pKa = 4.33QITARR304 pKa = 11.84YY305 pKa = 7.8PNSLWIAAYY314 pKa = 8.94PDD316 pKa = 3.74YY317 pKa = 10.64EE318 pKa = 4.52VRR320 pKa = 11.84PEE322 pKa = 4.38PYY324 pKa = 9.07WGVYY328 pKa = 10.28PNMEE332 pKa = 4.23HH333 pKa = 5.47TRR335 pKa = 11.84WWQFTSTGLAGGLDD349 pKa = 3.76KK350 pKa = 11.39NVVIINDD357 pKa = 3.56GDD359 pKa = 4.01SLVNKK364 pKa = 9.9EE365 pKa = 4.19KK366 pKa = 11.01EE367 pKa = 4.39EE368 pKa = 4.09EE369 pKa = 4.06NMDD372 pKa = 3.59YY373 pKa = 10.92VVRR376 pKa = 11.84SEE378 pKa = 4.67SGSQGYY384 pKa = 10.02VGVVNGRR391 pKa = 11.84VFGIGSMGTVDD402 pKa = 3.97ALRR405 pKa = 11.84SAGAKK410 pKa = 10.07HH411 pKa = 5.78LTLPDD416 pKa = 3.46ADD418 pKa = 3.99FDD420 pKa = 4.53RR421 pKa = 11.84FLNSQSNDD429 pKa = 3.02TQAVAKK435 pKa = 10.01AVSEE439 pKa = 4.22ASASVVKK446 pKa = 10.68AIEE449 pKa = 3.89EE450 pKa = 4.05RR451 pKa = 11.84AQATQGQTGVV461 pKa = 3.31
Molecular weight: 50.48 kDa
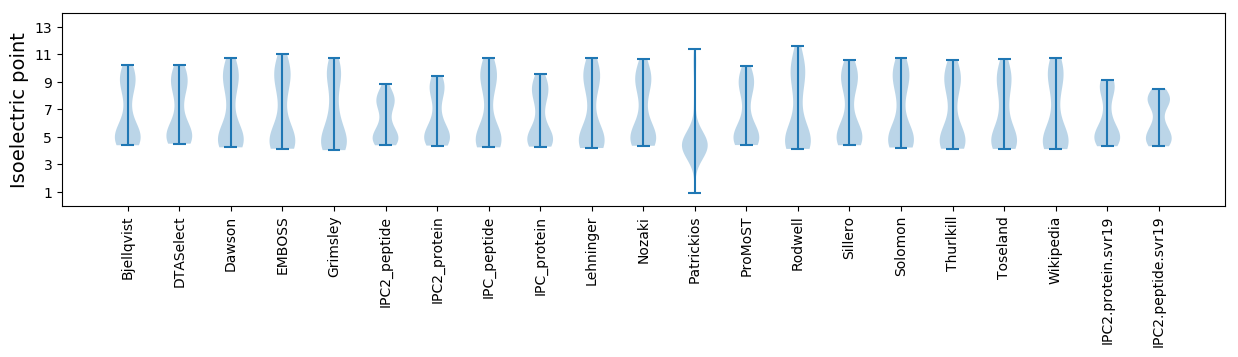
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D6BEX6|A0A4D6BEX6_9CAUD Uncharacterized protein OS=Streptococcus phage Javan536 OX=2548249 GN=Javan536_0014 PE=4 SV=1
MM1 pKa = 7.79IVIRR5 pKa = 11.84HH6 pKa = 4.96NAKK9 pKa = 10.45VIEE12 pKa = 4.43GQVAVLNGTQYY23 pKa = 11.38DD24 pKa = 3.86IVRR27 pKa = 11.84VSPNEE32 pKa = 3.62NFGLNRR38 pKa = 11.84YY39 pKa = 9.61DD40 pKa = 4.26FLTLRR45 pKa = 11.84KK46 pKa = 8.91HH47 pKa = 6.08KK48 pKa = 10.69KK49 pKa = 9.55VGG51 pKa = 3.25
MM1 pKa = 7.79IVIRR5 pKa = 11.84HH6 pKa = 4.96NAKK9 pKa = 10.45VIEE12 pKa = 4.43GQVAVLNGTQYY23 pKa = 11.38DD24 pKa = 3.86IVRR27 pKa = 11.84VSPNEE32 pKa = 3.62NFGLNRR38 pKa = 11.84YY39 pKa = 9.61DD40 pKa = 4.26FLTLRR45 pKa = 11.84KK46 pKa = 8.91HH47 pKa = 6.08KK48 pKa = 10.69KK49 pKa = 9.55VGG51 pKa = 3.25
Molecular weight: 5.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11581 |
47 |
1654 |
269.3 |
30.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.046 ± 0.596 | 0.518 ± 0.12 |
6.364 ± 0.255 | 6.606 ± 0.538 |
4.076 ± 0.191 | 7.193 ± 0.527 |
1.2 ± 0.121 | 5.725 ± 0.261 |
8.013 ± 0.491 | 7.745 ± 0.388 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.34 ± 0.187 | 6.398 ± 0.321 |
3.488 ± 0.415 | 4.404 ± 0.242 |
3.877 ± 0.305 | 6.407 ± 0.308 |
7.098 ± 0.396 | 6.39 ± 0.241 |
1.382 ± 0.153 | 3.73 ± 0.266 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
