
Streptococcus satellite phage Javan293
Taxonomy: Viruses; unclassified bacterial viruses
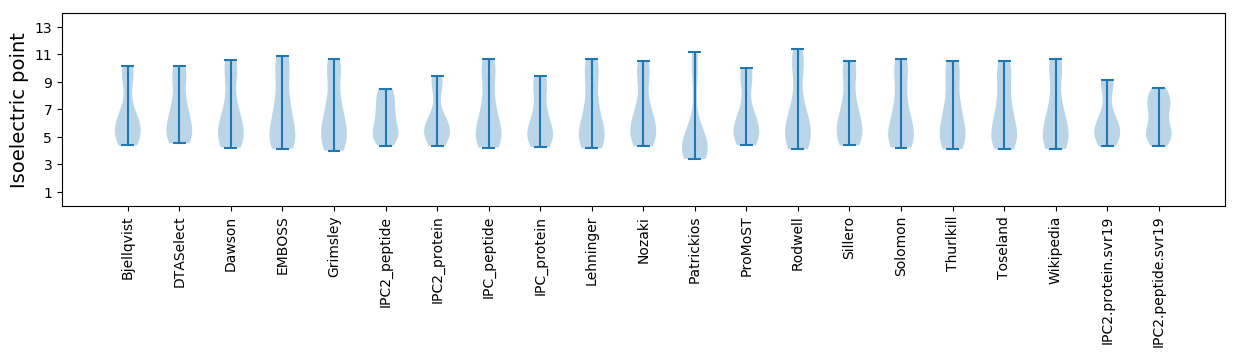
Average proteome isoelectric point is 6.39
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 22 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A4D5ZIV7|A0A4D5ZIV7_9VIRU Uncharacterized protein OS=Streptococcus satellite phage Javan293 OX=2558612 GN=JavanS293_0013 PE=4 SV=1
MM1 pKa = 7.39LVTYY5 pKa = 8.27PALFYY10 pKa = 11.3YY11 pKa = 10.68DD12 pKa = 4.09DD13 pKa = 4.06TNGTNVPYY21 pKa = 9.91FVHH24 pKa = 7.0FPDD27 pKa = 3.95IDD29 pKa = 3.78GSGTQGTSLSDD40 pKa = 3.47AMAMASEE47 pKa = 4.29YY48 pKa = 10.57LGIMVAYY55 pKa = 9.47YY56 pKa = 9.98IEE58 pKa = 4.7SNRR61 pKa = 11.84EE62 pKa = 3.61LPKK65 pKa = 10.48PSEE68 pKa = 4.16INNLSLMEE76 pKa = 4.53NNPFKK81 pKa = 10.86DD82 pKa = 3.62EE83 pKa = 4.59PDD85 pKa = 3.58FLSYY89 pKa = 11.23DD90 pKa = 3.51PGKK93 pKa = 10.83SFVSLVMVDD102 pKa = 3.19VAEE105 pKa = 4.16YY106 pKa = 10.54LGSQEE111 pKa = 4.64PIKK114 pKa = 10.26KK115 pKa = 8.15TLTIPRR121 pKa = 11.84WADD124 pKa = 3.04KK125 pKa = 10.79LGRR128 pKa = 11.84EE129 pKa = 4.06LGLNFSQTLTEE140 pKa = 5.14AIADD144 pKa = 3.94KK145 pKa = 10.63KK146 pKa = 10.1IQAA149 pKa = 4.07
MM1 pKa = 7.39LVTYY5 pKa = 8.27PALFYY10 pKa = 11.3YY11 pKa = 10.68DD12 pKa = 4.09DD13 pKa = 4.06TNGTNVPYY21 pKa = 9.91FVHH24 pKa = 7.0FPDD27 pKa = 3.95IDD29 pKa = 3.78GSGTQGTSLSDD40 pKa = 3.47AMAMASEE47 pKa = 4.29YY48 pKa = 10.57LGIMVAYY55 pKa = 9.47YY56 pKa = 9.98IEE58 pKa = 4.7SNRR61 pKa = 11.84EE62 pKa = 3.61LPKK65 pKa = 10.48PSEE68 pKa = 4.16INNLSLMEE76 pKa = 4.53NNPFKK81 pKa = 10.86DD82 pKa = 3.62EE83 pKa = 4.59PDD85 pKa = 3.58FLSYY89 pKa = 11.23DD90 pKa = 3.51PGKK93 pKa = 10.83SFVSLVMVDD102 pKa = 3.19VAEE105 pKa = 4.16YY106 pKa = 10.54LGSQEE111 pKa = 4.64PIKK114 pKa = 10.26KK115 pKa = 8.15TLTIPRR121 pKa = 11.84WADD124 pKa = 3.04KK125 pKa = 10.79LGRR128 pKa = 11.84EE129 pKa = 4.06LGLNFSQTLTEE140 pKa = 5.14AIADD144 pKa = 3.94KK145 pKa = 10.63KK146 pKa = 10.1IQAA149 pKa = 4.07
Molecular weight: 16.65 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D5ZJD1|A0A4D5ZJD1_9VIRU Mobile element protein OS=Streptococcus satellite phage Javan293 OX=2558612 GN=JavanS293_0009 PE=4 SV=1
MM1 pKa = 7.38IMKK4 pKa = 8.9ITEE7 pKa = 4.09VKK9 pKa = 10.48KK10 pKa = 10.14KK11 pKa = 10.37DD12 pKa = 3.24GSIVYY17 pKa = 9.61RR18 pKa = 11.84ANIYY22 pKa = 10.71LGVDD26 pKa = 3.6NITGKK31 pKa = 10.25KK32 pKa = 9.88VKK34 pKa = 9.83TSVTGRR40 pKa = 11.84TKK42 pKa = 10.86KK43 pKa = 10.07EE44 pKa = 3.87VSRR47 pKa = 11.84KK48 pKa = 9.1AQNAQLDD55 pKa = 3.98FKK57 pKa = 11.24INGSTVYY64 pKa = 9.03KK65 pKa = 9.41TVQIEE70 pKa = 4.63SYY72 pKa = 10.83KK73 pKa = 10.62EE74 pKa = 3.74LAEE77 pKa = 4.3LWLEE81 pKa = 4.29SYY83 pKa = 10.96KK84 pKa = 10.44LTVKK88 pKa = 10.09PQTYY92 pKa = 10.46RR93 pKa = 11.84NTTLFLKK100 pKa = 10.7NHH102 pKa = 7.14LLPMFGDD109 pKa = 3.45MRR111 pKa = 11.84LEE113 pKa = 4.46KK114 pKa = 9.33ITPHH118 pKa = 5.27IVQRR122 pKa = 11.84FINRR126 pKa = 11.84LAVTLANFRR135 pKa = 11.84VPVSINSRR143 pKa = 11.84ILQYY147 pKa = 11.38AVTLQLIPYY156 pKa = 8.72NPARR160 pKa = 11.84DD161 pKa = 3.78VILPPKK167 pKa = 8.79PAKK170 pKa = 10.29DD171 pKa = 3.41PQKK174 pKa = 9.74ITFIDD179 pKa = 3.62KK180 pKa = 10.63QDD182 pKa = 3.55LKK184 pKa = 11.4KK185 pKa = 11.04LLDD188 pKa = 3.84YY189 pKa = 11.1VEE191 pKa = 5.15KK192 pKa = 10.71ISIQHH197 pKa = 4.0YY198 pKa = 8.32TGYY201 pKa = 10.97FDD203 pKa = 3.53MVLYY207 pKa = 8.48KK208 pKa = 10.31TLLATGCRR216 pKa = 11.84IGEE219 pKa = 4.05LLALNWSDD227 pKa = 3.85IDD229 pKa = 4.27FKK231 pKa = 11.51EE232 pKa = 3.89KK233 pKa = 9.84TLNISKK239 pKa = 9.19TYY241 pKa = 9.02NGEE244 pKa = 3.65MGLIGTTKK252 pKa = 10.33SKK254 pKa = 10.91AGNRR258 pKa = 11.84TISVDD263 pKa = 3.53TNTLNMLRR271 pKa = 11.84LYY273 pKa = 10.34NARR276 pKa = 11.84QRR278 pKa = 11.84QMFSEE283 pKa = 4.43IGAPRR288 pKa = 11.84PSVVFSAPTRR298 pKa = 11.84LYY300 pKa = 10.41QLRR303 pKa = 11.84TTIQRR308 pKa = 11.84RR309 pKa = 11.84LDD311 pKa = 4.24DD312 pKa = 3.92YY313 pKa = 11.32CHH315 pKa = 6.78TIDD318 pKa = 3.84IPRR321 pKa = 11.84FTLHH325 pKa = 7.29AFRR328 pKa = 11.84HH329 pKa = 4.72THH331 pKa = 7.12ASLLLNAGISYY342 pKa = 10.68KK343 pKa = 10.23EE344 pKa = 3.61LQYY347 pKa = 11.51RR348 pKa = 11.84LGHH351 pKa = 6.41ANISMTLDD359 pKa = 3.58VYY361 pKa = 11.5SHH363 pKa = 7.06LSKK366 pKa = 11.04DD367 pKa = 3.52KK368 pKa = 10.43EE369 pKa = 4.37KK370 pKa = 11.05EE371 pKa = 3.74AVTYY375 pKa = 8.83YY376 pKa = 10.86EE377 pKa = 4.2KK378 pKa = 10.84AINNLL383 pKa = 3.4
MM1 pKa = 7.38IMKK4 pKa = 8.9ITEE7 pKa = 4.09VKK9 pKa = 10.48KK10 pKa = 10.14KK11 pKa = 10.37DD12 pKa = 3.24GSIVYY17 pKa = 9.61RR18 pKa = 11.84ANIYY22 pKa = 10.71LGVDD26 pKa = 3.6NITGKK31 pKa = 10.25KK32 pKa = 9.88VKK34 pKa = 9.83TSVTGRR40 pKa = 11.84TKK42 pKa = 10.86KK43 pKa = 10.07EE44 pKa = 3.87VSRR47 pKa = 11.84KK48 pKa = 9.1AQNAQLDD55 pKa = 3.98FKK57 pKa = 11.24INGSTVYY64 pKa = 9.03KK65 pKa = 9.41TVQIEE70 pKa = 4.63SYY72 pKa = 10.83KK73 pKa = 10.62EE74 pKa = 3.74LAEE77 pKa = 4.3LWLEE81 pKa = 4.29SYY83 pKa = 10.96KK84 pKa = 10.44LTVKK88 pKa = 10.09PQTYY92 pKa = 10.46RR93 pKa = 11.84NTTLFLKK100 pKa = 10.7NHH102 pKa = 7.14LLPMFGDD109 pKa = 3.45MRR111 pKa = 11.84LEE113 pKa = 4.46KK114 pKa = 9.33ITPHH118 pKa = 5.27IVQRR122 pKa = 11.84FINRR126 pKa = 11.84LAVTLANFRR135 pKa = 11.84VPVSINSRR143 pKa = 11.84ILQYY147 pKa = 11.38AVTLQLIPYY156 pKa = 8.72NPARR160 pKa = 11.84DD161 pKa = 3.78VILPPKK167 pKa = 8.79PAKK170 pKa = 10.29DD171 pKa = 3.41PQKK174 pKa = 9.74ITFIDD179 pKa = 3.62KK180 pKa = 10.63QDD182 pKa = 3.55LKK184 pKa = 11.4KK185 pKa = 11.04LLDD188 pKa = 3.84YY189 pKa = 11.1VEE191 pKa = 5.15KK192 pKa = 10.71ISIQHH197 pKa = 4.0YY198 pKa = 8.32TGYY201 pKa = 10.97FDD203 pKa = 3.53MVLYY207 pKa = 8.48KK208 pKa = 10.31TLLATGCRR216 pKa = 11.84IGEE219 pKa = 4.05LLALNWSDD227 pKa = 3.85IDD229 pKa = 4.27FKK231 pKa = 11.51EE232 pKa = 3.89KK233 pKa = 9.84TLNISKK239 pKa = 9.19TYY241 pKa = 9.02NGEE244 pKa = 3.65MGLIGTTKK252 pKa = 10.33SKK254 pKa = 10.91AGNRR258 pKa = 11.84TISVDD263 pKa = 3.53TNTLNMLRR271 pKa = 11.84LYY273 pKa = 10.34NARR276 pKa = 11.84QRR278 pKa = 11.84QMFSEE283 pKa = 4.43IGAPRR288 pKa = 11.84PSVVFSAPTRR298 pKa = 11.84LYY300 pKa = 10.41QLRR303 pKa = 11.84TTIQRR308 pKa = 11.84RR309 pKa = 11.84LDD311 pKa = 4.24DD312 pKa = 3.92YY313 pKa = 11.32CHH315 pKa = 6.78TIDD318 pKa = 3.84IPRR321 pKa = 11.84FTLHH325 pKa = 7.29AFRR328 pKa = 11.84HH329 pKa = 4.72THH331 pKa = 7.12ASLLLNAGISYY342 pKa = 10.68KK343 pKa = 10.23EE344 pKa = 3.61LQYY347 pKa = 11.51RR348 pKa = 11.84LGHH351 pKa = 6.41ANISMTLDD359 pKa = 3.58VYY361 pKa = 11.5SHH363 pKa = 7.06LSKK366 pKa = 11.04DD367 pKa = 3.52KK368 pKa = 10.43EE369 pKa = 4.37KK370 pKa = 11.05EE371 pKa = 3.74AVTYY375 pKa = 8.83YY376 pKa = 10.86EE377 pKa = 4.2KK378 pKa = 10.84AINNLL383 pKa = 3.4
Molecular weight: 44.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3805 |
59 |
474 |
173.0 |
19.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.938 ± 0.416 | 0.42 ± 0.139 |
5.834 ± 0.368 | 7.963 ± 0.698 |
4.074 ± 0.382 | 4.625 ± 0.471 |
2.208 ± 0.242 | 7.201 ± 0.324 |
8.673 ± 0.407 | 10.145 ± 0.567 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.024 ± 0.263 | 4.967 ± 0.361 |
3.259 ± 0.337 | 4.625 ± 0.272 |
5.151 ± 0.384 | 5.02 ± 0.316 |
5.887 ± 0.515 | 5.466 ± 0.334 |
0.841 ± 0.156 | 4.678 ± 0.3 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
