
Actibacterium mucosum KCTC 23349
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Actibacterium; Actibacterium mucosum
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins
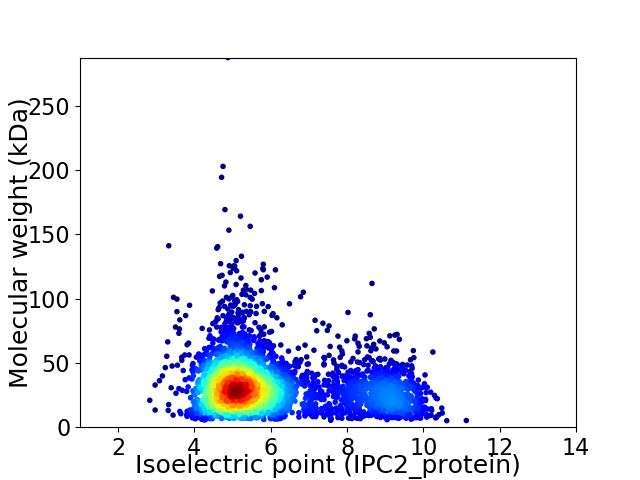
Virtual 2D-PAGE plot for 3616 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A037ZFG2|A0A037ZFG2_9RHOB Phosphonates import ATP-binding protein PhnC OS=Actibacterium mucosum KCTC 23349 OX=1454373 GN=phnC PE=3 SV=1
MM1 pKa = 7.78KK2 pKa = 9.82STTFTAKK9 pKa = 10.29RR10 pKa = 11.84AGPEE14 pKa = 3.81GARR17 pKa = 11.84TNEE20 pKa = 3.45VMFRR24 pKa = 11.84DD25 pKa = 4.26GGLDD29 pKa = 3.38PLNAFSPLLPVDD41 pKa = 5.21DD42 pKa = 5.44LFANQWYY49 pKa = 9.85LLNTGQSGGVPGVDD63 pKa = 3.53LNVVDD68 pKa = 4.37VWDD71 pKa = 5.18DD72 pKa = 3.56YY73 pKa = 10.85TGEE76 pKa = 4.2GVTVGIWDD84 pKa = 4.62DD85 pKa = 3.74GVQYY89 pKa = 9.02THH91 pKa = 7.41HH92 pKa = 7.5DD93 pKa = 3.44LDD95 pKa = 4.77DD96 pKa = 4.8NYY98 pKa = 11.56DD99 pKa = 3.86EE100 pKa = 4.86NLHH103 pKa = 5.49ITVGGFVHH111 pKa = 7.36DD112 pKa = 4.75PAPQAFDD119 pKa = 3.7SAHH122 pKa = 5.21GTSVAGIIGAEE133 pKa = 4.1NNGEE137 pKa = 4.18GTVGVAYY144 pKa = 10.09DD145 pKa = 3.47ATLAGVDD152 pKa = 3.18IFFDD156 pKa = 4.08FFLNFEE162 pKa = 4.03QSFYY166 pKa = 11.27EE167 pKa = 4.23LDD169 pKa = 3.59NFDD172 pKa = 3.96VTNHH176 pKa = 4.32SWGWSSPYY184 pKa = 10.24LDD186 pKa = 4.91SIYY189 pKa = 10.11DD190 pKa = 3.62TSGSSGTDD198 pKa = 2.64WQTFFGGIQQSVITGRR214 pKa = 11.84DD215 pKa = 3.18GLGTINVVANGNDD228 pKa = 3.46RR229 pKa = 11.84EE230 pKa = 4.18IGRR233 pKa = 11.84DD234 pKa = 3.73GNDD237 pKa = 3.19SNLNNIPQTIAVGATSHH254 pKa = 6.35EE255 pKa = 4.6GFVSYY260 pKa = 11.12YY261 pKa = 8.42STPGANLLISAPSNGEE277 pKa = 3.93PGSGVYY283 pKa = 7.51TTDD286 pKa = 2.93RR287 pKa = 11.84TGFSGYY293 pKa = 10.24EE294 pKa = 3.81LGDD297 pKa = 3.56YY298 pKa = 10.44TSDD301 pKa = 3.76FGGTSSAAPAVAGVVALMLEE321 pKa = 4.43ANPDD325 pKa = 3.7LGWRR329 pKa = 11.84DD330 pKa = 3.49VQEE333 pKa = 4.17ILAVTARR340 pKa = 11.84HH341 pKa = 5.66TGSDD345 pKa = 2.96IGAGPSFEE353 pKa = 5.14EE354 pKa = 3.8LHH356 pKa = 5.27TWEE359 pKa = 5.4FNGATNWNGGGLHH372 pKa = 6.85FSNDD376 pKa = 3.45YY377 pKa = 11.18GFGLIDD383 pKa = 3.58ALAAVRR389 pKa = 11.84LAEE392 pKa = 4.17TWTEE396 pKa = 4.07QQTSANWEE404 pKa = 4.14TPIVGSQTLNTFIQDD419 pKa = 3.58GNPNGISFTFEE430 pKa = 3.41VTNAFDD436 pKa = 4.08LEE438 pKa = 4.67HH439 pKa = 6.86VGLTLDD445 pKa = 4.07FASGRR450 pKa = 11.84TGDD453 pKa = 3.65YY454 pKa = 10.43TIVLVSPDD462 pKa = 3.39GTRR465 pKa = 11.84STLAVSNTGDD475 pKa = 3.36AFTDD479 pKa = 2.88EE480 pKa = 4.4WFYY483 pKa = 10.9MSNEE487 pKa = 3.71FRR489 pKa = 11.84GEE491 pKa = 4.18TGVGTWTVEE500 pKa = 3.99IVDD503 pKa = 3.79EE504 pKa = 4.22RR505 pKa = 11.84LGVSGILEE513 pKa = 4.06SAEE516 pKa = 3.78LQFFGHH522 pKa = 6.31SADD525 pKa = 4.29ANDD528 pKa = 3.71TYY530 pKa = 11.62VYY532 pKa = 10.19TNEE535 pKa = 4.07FSDD538 pKa = 3.98FAGDD542 pKa = 3.93GNHH545 pKa = 5.2VTVLQDD551 pKa = 3.25TNGGVDD557 pKa = 3.73TLNAAAVTADD567 pKa = 3.42AFVFLNFGAFIDD579 pKa = 3.83STFISGVTGIEE590 pKa = 3.74NVYY593 pKa = 10.02TGDD596 pKa = 3.35GNDD599 pKa = 2.97RR600 pKa = 11.84VFGNNDD606 pKa = 3.82DD607 pKa = 3.74NTLWGGRR614 pKa = 11.84GNDD617 pKa = 3.91QISGGQGSDD626 pKa = 3.26TLVDD630 pKa = 3.65GGGTDD635 pKa = 3.59VLIGGAGGDD644 pKa = 3.5IYY646 pKa = 11.58SLGVDD651 pKa = 3.41TAFDD655 pKa = 4.24IIFGFADD662 pKa = 3.91GFDD665 pKa = 4.95FIQFEE670 pKa = 4.5AEE672 pKa = 4.0DD673 pKa = 3.7VSYY676 pKa = 11.61EE677 pKa = 4.0DD678 pKa = 5.31LSVFNLNAGRR688 pKa = 11.84VLVQYY693 pKa = 10.2QGEE696 pKa = 4.47SVLVIEE702 pKa = 5.49AGLTAADD709 pKa = 3.66FDD711 pKa = 4.17EE712 pKa = 6.56ADD714 pKa = 3.65FLFAA718 pKa = 6.2
MM1 pKa = 7.78KK2 pKa = 9.82STTFTAKK9 pKa = 10.29RR10 pKa = 11.84AGPEE14 pKa = 3.81GARR17 pKa = 11.84TNEE20 pKa = 3.45VMFRR24 pKa = 11.84DD25 pKa = 4.26GGLDD29 pKa = 3.38PLNAFSPLLPVDD41 pKa = 5.21DD42 pKa = 5.44LFANQWYY49 pKa = 9.85LLNTGQSGGVPGVDD63 pKa = 3.53LNVVDD68 pKa = 4.37VWDD71 pKa = 5.18DD72 pKa = 3.56YY73 pKa = 10.85TGEE76 pKa = 4.2GVTVGIWDD84 pKa = 4.62DD85 pKa = 3.74GVQYY89 pKa = 9.02THH91 pKa = 7.41HH92 pKa = 7.5DD93 pKa = 3.44LDD95 pKa = 4.77DD96 pKa = 4.8NYY98 pKa = 11.56DD99 pKa = 3.86EE100 pKa = 4.86NLHH103 pKa = 5.49ITVGGFVHH111 pKa = 7.36DD112 pKa = 4.75PAPQAFDD119 pKa = 3.7SAHH122 pKa = 5.21GTSVAGIIGAEE133 pKa = 4.1NNGEE137 pKa = 4.18GTVGVAYY144 pKa = 10.09DD145 pKa = 3.47ATLAGVDD152 pKa = 3.18IFFDD156 pKa = 4.08FFLNFEE162 pKa = 4.03QSFYY166 pKa = 11.27EE167 pKa = 4.23LDD169 pKa = 3.59NFDD172 pKa = 3.96VTNHH176 pKa = 4.32SWGWSSPYY184 pKa = 10.24LDD186 pKa = 4.91SIYY189 pKa = 10.11DD190 pKa = 3.62TSGSSGTDD198 pKa = 2.64WQTFFGGIQQSVITGRR214 pKa = 11.84DD215 pKa = 3.18GLGTINVVANGNDD228 pKa = 3.46RR229 pKa = 11.84EE230 pKa = 4.18IGRR233 pKa = 11.84DD234 pKa = 3.73GNDD237 pKa = 3.19SNLNNIPQTIAVGATSHH254 pKa = 6.35EE255 pKa = 4.6GFVSYY260 pKa = 11.12YY261 pKa = 8.42STPGANLLISAPSNGEE277 pKa = 3.93PGSGVYY283 pKa = 7.51TTDD286 pKa = 2.93RR287 pKa = 11.84TGFSGYY293 pKa = 10.24EE294 pKa = 3.81LGDD297 pKa = 3.56YY298 pKa = 10.44TSDD301 pKa = 3.76FGGTSSAAPAVAGVVALMLEE321 pKa = 4.43ANPDD325 pKa = 3.7LGWRR329 pKa = 11.84DD330 pKa = 3.49VQEE333 pKa = 4.17ILAVTARR340 pKa = 11.84HH341 pKa = 5.66TGSDD345 pKa = 2.96IGAGPSFEE353 pKa = 5.14EE354 pKa = 3.8LHH356 pKa = 5.27TWEE359 pKa = 5.4FNGATNWNGGGLHH372 pKa = 6.85FSNDD376 pKa = 3.45YY377 pKa = 11.18GFGLIDD383 pKa = 3.58ALAAVRR389 pKa = 11.84LAEE392 pKa = 4.17TWTEE396 pKa = 4.07QQTSANWEE404 pKa = 4.14TPIVGSQTLNTFIQDD419 pKa = 3.58GNPNGISFTFEE430 pKa = 3.41VTNAFDD436 pKa = 4.08LEE438 pKa = 4.67HH439 pKa = 6.86VGLTLDD445 pKa = 4.07FASGRR450 pKa = 11.84TGDD453 pKa = 3.65YY454 pKa = 10.43TIVLVSPDD462 pKa = 3.39GTRR465 pKa = 11.84STLAVSNTGDD475 pKa = 3.36AFTDD479 pKa = 2.88EE480 pKa = 4.4WFYY483 pKa = 10.9MSNEE487 pKa = 3.71FRR489 pKa = 11.84GEE491 pKa = 4.18TGVGTWTVEE500 pKa = 3.99IVDD503 pKa = 3.79EE504 pKa = 4.22RR505 pKa = 11.84LGVSGILEE513 pKa = 4.06SAEE516 pKa = 3.78LQFFGHH522 pKa = 6.31SADD525 pKa = 4.29ANDD528 pKa = 3.71TYY530 pKa = 11.62VYY532 pKa = 10.19TNEE535 pKa = 4.07FSDD538 pKa = 3.98FAGDD542 pKa = 3.93GNHH545 pKa = 5.2VTVLQDD551 pKa = 3.25TNGGVDD557 pKa = 3.73TLNAAAVTADD567 pKa = 3.42AFVFLNFGAFIDD579 pKa = 3.83STFISGVTGIEE590 pKa = 3.74NVYY593 pKa = 10.02TGDD596 pKa = 3.35GNDD599 pKa = 2.97RR600 pKa = 11.84VFGNNDD606 pKa = 3.82DD607 pKa = 3.74NTLWGGRR614 pKa = 11.84GNDD617 pKa = 3.91QISGGQGSDD626 pKa = 3.26TLVDD630 pKa = 3.65GGGTDD635 pKa = 3.59VLIGGAGGDD644 pKa = 3.5IYY646 pKa = 11.58SLGVDD651 pKa = 3.41TAFDD655 pKa = 4.24IIFGFADD662 pKa = 3.91GFDD665 pKa = 4.95FIQFEE670 pKa = 4.5AEE672 pKa = 4.0DD673 pKa = 3.7VSYY676 pKa = 11.61EE677 pKa = 4.0DD678 pKa = 5.31LSVFNLNAGRR688 pKa = 11.84VLVQYY693 pKa = 10.2QGEE696 pKa = 4.47SVLVIEE702 pKa = 5.49AGLTAADD709 pKa = 3.66FDD711 pKa = 4.17EE712 pKa = 6.56ADD714 pKa = 3.65FLFAA718 pKa = 6.2
Molecular weight: 76.39 kDa
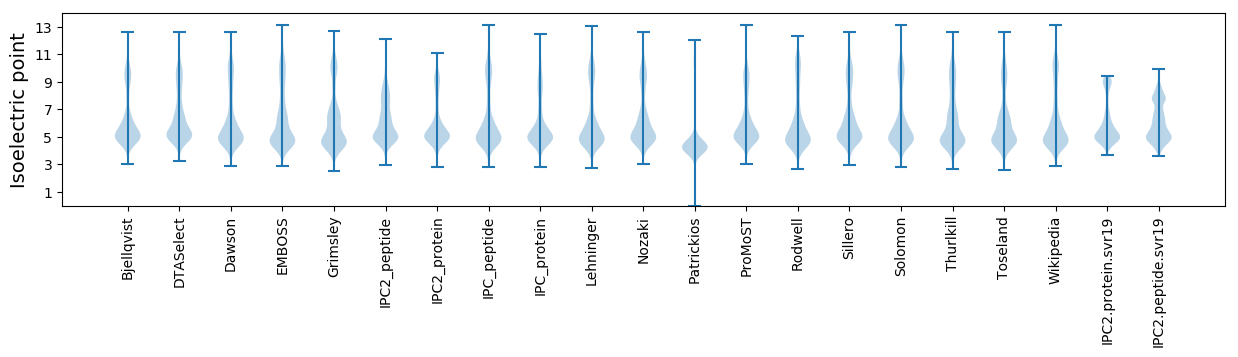
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A037ZGX6|A0A037ZGX6_9RHOB NAD(P) transhydrogenase subunit alpha OS=Actibacterium mucosum KCTC 23349 OX=1454373 GN=ACMU_13570 PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.5ILNARR34 pKa = 11.84RR35 pKa = 11.84LRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.5ILNARR34 pKa = 11.84RR35 pKa = 11.84LRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1123337 |
41 |
2766 |
310.7 |
33.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.5 ± 0.059 | 0.844 ± 0.013 |
6.222 ± 0.039 | 5.597 ± 0.04 |
3.91 ± 0.026 | 8.708 ± 0.035 |
2.055 ± 0.019 | 5.094 ± 0.03 |
3.143 ± 0.032 | 10.016 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.786 ± 0.02 | 2.852 ± 0.019 |
5.057 ± 0.028 | 3.412 ± 0.027 |
6.289 ± 0.038 | 4.959 ± 0.027 |
5.541 ± 0.025 | 7.465 ± 0.033 |
1.377 ± 0.015 | 2.174 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
