
Leucoagaricus sp. SymC.cos
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetidae; Agaricales; Agaricaceae; Leucoagaricus; unclassified Leucoagaricus
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
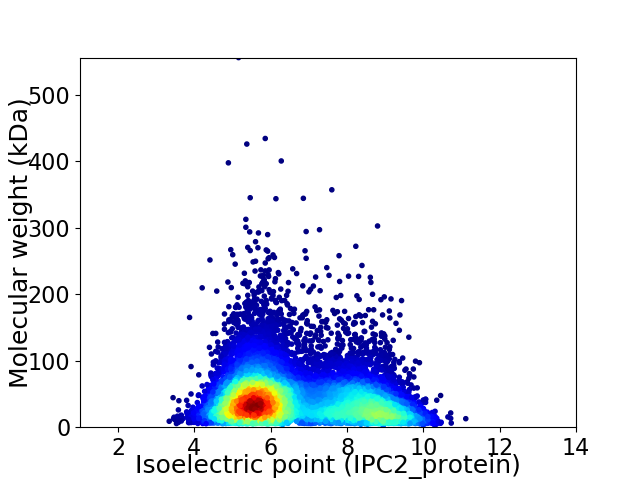
Virtual 2D-PAGE plot for 12747 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A137QRF3|A0A137QRF3_9AGAR Vacuolar amino acid transporter 1 OS=Leucoagaricus sp. SymC.cos OX=1714833 GN=AN958_04869 PE=3 SV=1
MM1 pKa = 7.47NSFFKK6 pKa = 10.58TLIALSLLAPGHH18 pKa = 6.92LFTLAGPVKK27 pKa = 10.23RR28 pKa = 11.84QDD30 pKa = 4.04DD31 pKa = 5.05GISTDD36 pKa = 3.55ATSTTPLASSQLASSTPTFPSDD58 pKa = 2.99SDD60 pKa = 3.83TFSVPTASPSDD71 pKa = 4.03FPSDD75 pKa = 3.53PTSTPSCSCQMTDD88 pKa = 3.42MNPIPVFSSGTPVDD102 pKa = 4.25PPAPTDD108 pKa = 3.34TMIPTWSDD116 pKa = 2.98GSHH119 pKa = 5.93CHH121 pKa = 5.68HH122 pKa = 7.2HH123 pKa = 6.86GGMPNPSDD131 pKa = 3.95GFPGGLGGDD140 pKa = 3.49NSTPTGDD147 pKa = 3.0IPYY150 pKa = 9.37PGGAMPTPPSTLTAVSVPFPSDD172 pKa = 3.22TSPAPPSTATEE183 pKa = 4.03VGVPAFSDD191 pKa = 3.66SSAEE195 pKa = 3.94PTPLSTATEE204 pKa = 4.16VGTPYY209 pKa = 10.57PGDD212 pKa = 3.89ASPTPPSTLTAVSITDD228 pKa = 3.53PTDD231 pKa = 3.23VPEE234 pKa = 4.28TSGGEE239 pKa = 3.99STAIPTSSTVTDD251 pKa = 4.16SEE253 pKa = 4.86DD254 pKa = 3.46PAPTSTT260 pKa = 4.5
MM1 pKa = 7.47NSFFKK6 pKa = 10.58TLIALSLLAPGHH18 pKa = 6.92LFTLAGPVKK27 pKa = 10.23RR28 pKa = 11.84QDD30 pKa = 4.04DD31 pKa = 5.05GISTDD36 pKa = 3.55ATSTTPLASSQLASSTPTFPSDD58 pKa = 2.99SDD60 pKa = 3.83TFSVPTASPSDD71 pKa = 4.03FPSDD75 pKa = 3.53PTSTPSCSCQMTDD88 pKa = 3.42MNPIPVFSSGTPVDD102 pKa = 4.25PPAPTDD108 pKa = 3.34TMIPTWSDD116 pKa = 2.98GSHH119 pKa = 5.93CHH121 pKa = 5.68HH122 pKa = 7.2HH123 pKa = 6.86GGMPNPSDD131 pKa = 3.95GFPGGLGGDD140 pKa = 3.49NSTPTGDD147 pKa = 3.0IPYY150 pKa = 9.37PGGAMPTPPSTLTAVSVPFPSDD172 pKa = 3.22TSPAPPSTATEE183 pKa = 4.03VGVPAFSDD191 pKa = 3.66SSAEE195 pKa = 3.94PTPLSTATEE204 pKa = 4.16VGTPYY209 pKa = 10.57PGDD212 pKa = 3.89ASPTPPSTLTAVSITDD228 pKa = 3.53PTDD231 pKa = 3.23VPEE234 pKa = 4.28TSGGEE239 pKa = 3.99STAIPTSSTVTDD251 pKa = 4.16SEE253 pKa = 4.86DD254 pKa = 3.46PAPTSTT260 pKa = 4.5
Molecular weight: 26.05 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A137QIR8|A0A137QIR8_9AGAR von Willebrand factor A domain-containing protein 8 OS=Leucoagaricus sp. SymC.cos OX=1714833 GN=AN958_09143 PE=4 SV=1
MM1 pKa = 7.79TIRR4 pKa = 11.84PRR6 pKa = 11.84PRR8 pKa = 11.84TGTGMVYY15 pKa = 10.32RR16 pKa = 11.84KK17 pKa = 10.01SNSGSFTPNLAGIGAGPGPGPSAGVGTGGSRR48 pKa = 11.84IRR50 pKa = 11.84APMPLATASFTSVTMNTGSGGSVGSVVGKK79 pKa = 10.04SGTMRR84 pKa = 11.84NVSSGLAVRR93 pKa = 11.84RR94 pKa = 11.84ARR96 pKa = 11.84TPGPRR101 pKa = 11.84SASSPSPSPSSSSMPPPPSSMSATPIGQAFF131 pKa = 3.41
MM1 pKa = 7.79TIRR4 pKa = 11.84PRR6 pKa = 11.84PRR8 pKa = 11.84TGTGMVYY15 pKa = 10.32RR16 pKa = 11.84KK17 pKa = 10.01SNSGSFTPNLAGIGAGPGPGPSAGVGTGGSRR48 pKa = 11.84IRR50 pKa = 11.84APMPLATASFTSVTMNTGSGGSVGSVVGKK79 pKa = 10.04SGTMRR84 pKa = 11.84NVSSGLAVRR93 pKa = 11.84RR94 pKa = 11.84ARR96 pKa = 11.84TPGPRR101 pKa = 11.84SASSPSPSPSSSSMPPPPSSMSATPIGQAFF131 pKa = 3.41
Molecular weight: 12.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
5393887 |
16 |
4977 |
423.1 |
46.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.709 ± 0.019 | 1.268 ± 0.01 |
5.454 ± 0.014 | 5.904 ± 0.021 |
3.91 ± 0.016 | 6.396 ± 0.025 |
2.586 ± 0.01 | 5.262 ± 0.017 |
4.851 ± 0.019 | 9.346 ± 0.022 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.042 ± 0.008 | 3.791 ± 0.012 |
6.376 ± 0.03 | 3.838 ± 0.013 |
5.85 ± 0.017 | 8.596 ± 0.032 |
6.089 ± 0.016 | 6.316 ± 0.016 |
1.462 ± 0.008 | 2.745 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
