
Lachnospiraceae bacterium RM5
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; unclassified Lachnospiraceae
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
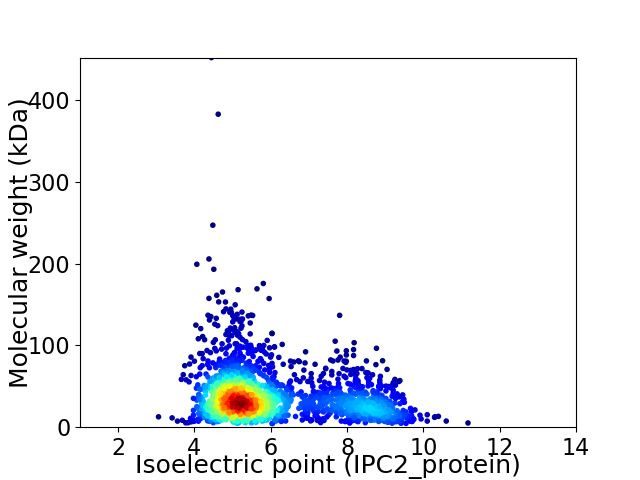
Virtual 2D-PAGE plot for 1991 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H9AAY1|A0A1H9AAY1_9FIRM Protease PrsW OS=Lachnospiraceae bacterium RM5 OX=1200721 GN=SAMN02910289_00492 PE=3 SV=1
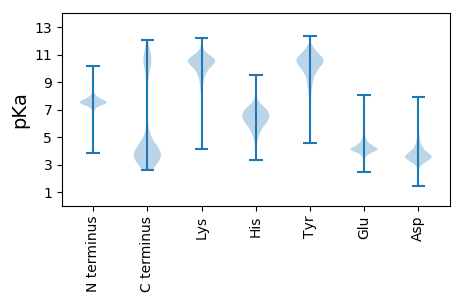
MM1 pKa = 7.91SYY3 pKa = 10.93RR4 pKa = 11.84DD5 pKa = 3.48DD6 pKa = 3.63RR7 pKa = 11.84RR8 pKa = 11.84NKK10 pKa = 8.39EE11 pKa = 4.08TEE13 pKa = 3.55KK14 pKa = 11.23DD15 pKa = 3.33DD16 pKa = 4.41FFIEE20 pKa = 5.74FYY22 pKa = 11.29DD23 pKa = 4.43NDD25 pKa = 3.89YY26 pKa = 11.83DD27 pKa = 6.63DD28 pKa = 5.6IDD30 pKa = 4.1EE31 pKa = 4.73KK32 pKa = 11.56NSDD35 pKa = 4.11EE36 pKa = 5.17EE37 pKa = 3.91FDD39 pKa = 4.35EE40 pKa = 4.73YY41 pKa = 11.49DD42 pKa = 4.04DD43 pKa = 4.16YY44 pKa = 12.03EE45 pKa = 4.42EE46 pKa = 5.27YY47 pKa = 10.79EE48 pKa = 4.21DD49 pKa = 4.32DD50 pKa = 4.33EE51 pKa = 4.96EE52 pKa = 4.92YY53 pKa = 11.08EE54 pKa = 4.21EE55 pKa = 4.65YY56 pKa = 10.68EE57 pKa = 3.94GDD59 pKa = 3.84EE60 pKa = 4.4EE61 pKa = 4.7YY62 pKa = 11.1EE63 pKa = 4.35EE64 pKa = 4.64YY65 pKa = 10.94DD66 pKa = 3.6EE67 pKa = 6.6LDD69 pKa = 4.34DD70 pKa = 5.41EE71 pKa = 6.37DD72 pKa = 6.25YY73 pKa = 11.69DD74 pKa = 5.83DD75 pKa = 6.79DD76 pKa = 6.0YY77 pKa = 12.01DD78 pKa = 6.1DD79 pKa = 6.01YY80 pKa = 12.07DD81 pKa = 4.24DD82 pKa = 4.2EE83 pKa = 5.9PYY85 pKa = 10.61VEE87 pKa = 5.43KK88 pKa = 10.0NTKK91 pKa = 8.72VKK93 pKa = 10.69KK94 pKa = 10.47KK95 pKa = 9.08LTSNQKK101 pKa = 8.83VNRR104 pKa = 11.84AIAILSGVIFCFTAFYY120 pKa = 10.38GAIAISSFAGKK131 pKa = 8.07TKK133 pKa = 10.38AAQEE137 pKa = 4.15APADD141 pKa = 3.83NQTEE145 pKa = 4.17AEE147 pKa = 4.11EE148 pKa = 4.87GFFVSQNSDD157 pKa = 2.59ATISQEE163 pKa = 5.02IKK165 pKa = 9.91DD166 pKa = 4.19TLNSATLEE174 pKa = 4.16PLTTGYY180 pKa = 10.42EE181 pKa = 4.22AIDD184 pKa = 3.44NSVANMLKK192 pKa = 8.65VTCKK196 pKa = 10.91DD197 pKa = 4.25DD198 pKa = 3.07MTTYY202 pKa = 11.17DD203 pKa = 3.82KK204 pKa = 11.32VRR206 pKa = 11.84NIYY209 pKa = 10.59DD210 pKa = 3.39YY211 pKa = 11.78MMFTYY216 pKa = 10.21EE217 pKa = 3.86NADD220 pKa = 3.13ASFVDD225 pKa = 4.0EE226 pKa = 4.58DD227 pKa = 4.62SVYY230 pKa = 11.14DD231 pKa = 3.56FCNKK235 pKa = 9.75YY236 pKa = 10.84SYY238 pKa = 11.11TNYY241 pKa = 9.89FDD243 pKa = 4.82MEE245 pKa = 4.1MLYY248 pKa = 10.56RR249 pKa = 11.84ADD251 pKa = 4.38KK252 pKa = 11.23LMASKK257 pKa = 10.85AGDD260 pKa = 3.46SRR262 pKa = 11.84DD263 pKa = 3.71YY264 pKa = 11.57GSTFAIALRR273 pKa = 11.84ALGLDD278 pKa = 3.67AYY280 pKa = 10.54YY281 pKa = 11.02VATSSDD287 pKa = 3.44TNYY290 pKa = 10.75LAGLDD295 pKa = 3.97TIKK298 pKa = 10.86GYY300 pKa = 8.01TVVVIDD306 pKa = 3.81DD307 pKa = 3.81EE308 pKa = 5.79KK309 pKa = 11.63YY310 pKa = 9.9VFDD313 pKa = 4.81VNAEE317 pKa = 4.46DD318 pKa = 4.82IEE320 pKa = 4.73STDD323 pKa = 3.3SEE325 pKa = 4.72AGVVYY330 pKa = 10.72SFFGKK335 pKa = 10.31DD336 pKa = 3.53FLDD339 pKa = 4.2LPEE342 pKa = 4.52QYY344 pKa = 10.03TLEE347 pKa = 4.29DD348 pKa = 3.53AEE350 pKa = 4.38ASIEE354 pKa = 4.0AFGNFGTLGEE364 pKa = 4.28FGISVVATTKK374 pKa = 10.37SGEE377 pKa = 4.17YY378 pKa = 10.07SSGAVSYY385 pKa = 10.94NSSGEE390 pKa = 4.0NLIEE394 pKa = 4.97ADD396 pKa = 4.53TIGIDD401 pKa = 3.46QGEE404 pKa = 4.38TIYY407 pKa = 10.87FKK409 pKa = 10.74GTVSGSEE416 pKa = 4.0TNTWKK421 pKa = 11.02LIADD425 pKa = 4.09VYY427 pKa = 9.96DD428 pKa = 4.04HH429 pKa = 6.45NWNYY433 pKa = 7.98VTEE436 pKa = 4.16ATLYY440 pKa = 11.15NEE442 pKa = 4.21THH444 pKa = 6.52SNRR447 pKa = 11.84TDD449 pKa = 3.66EE450 pKa = 4.13ITYY453 pKa = 10.12APSRR457 pKa = 11.84GGFIVIRR464 pKa = 11.84FIVTDD469 pKa = 3.73EE470 pKa = 3.96NGRR473 pKa = 11.84TCTVKK478 pKa = 10.62FIADD482 pKa = 3.57VAGDD486 pKa = 3.79DD487 pKa = 4.51YY488 pKa = 11.7YY489 pKa = 11.4FEE491 pKa = 5.71PEE493 pKa = 3.84TTTVYY498 pKa = 8.05EE499 pKa = 4.48TTTEE503 pKa = 4.19EE504 pKa = 4.6YY505 pKa = 7.81TTEE508 pKa = 3.85EE509 pKa = 4.12VTTIEE514 pKa = 3.98EE515 pKa = 4.44TTTVEE520 pKa = 3.76EE521 pKa = 4.51TTTVEE526 pKa = 4.02EE527 pKa = 4.45TTSQEE532 pKa = 3.84EE533 pKa = 4.36TTSQEE538 pKa = 3.9EE539 pKa = 4.35SSSQEE544 pKa = 3.72EE545 pKa = 4.25TSSQEE550 pKa = 3.9EE551 pKa = 4.46TSSQTEE557 pKa = 4.49PEE559 pKa = 4.53PQPQPQPP566 pKa = 3.37
MM1 pKa = 7.91SYY3 pKa = 10.93RR4 pKa = 11.84DD5 pKa = 3.48DD6 pKa = 3.63RR7 pKa = 11.84RR8 pKa = 11.84NKK10 pKa = 8.39EE11 pKa = 4.08TEE13 pKa = 3.55KK14 pKa = 11.23DD15 pKa = 3.33DD16 pKa = 4.41FFIEE20 pKa = 5.74FYY22 pKa = 11.29DD23 pKa = 4.43NDD25 pKa = 3.89YY26 pKa = 11.83DD27 pKa = 6.63DD28 pKa = 5.6IDD30 pKa = 4.1EE31 pKa = 4.73KK32 pKa = 11.56NSDD35 pKa = 4.11EE36 pKa = 5.17EE37 pKa = 3.91FDD39 pKa = 4.35EE40 pKa = 4.73YY41 pKa = 11.49DD42 pKa = 4.04DD43 pKa = 4.16YY44 pKa = 12.03EE45 pKa = 4.42EE46 pKa = 5.27YY47 pKa = 10.79EE48 pKa = 4.21DD49 pKa = 4.32DD50 pKa = 4.33EE51 pKa = 4.96EE52 pKa = 4.92YY53 pKa = 11.08EE54 pKa = 4.21EE55 pKa = 4.65YY56 pKa = 10.68EE57 pKa = 3.94GDD59 pKa = 3.84EE60 pKa = 4.4EE61 pKa = 4.7YY62 pKa = 11.1EE63 pKa = 4.35EE64 pKa = 4.64YY65 pKa = 10.94DD66 pKa = 3.6EE67 pKa = 6.6LDD69 pKa = 4.34DD70 pKa = 5.41EE71 pKa = 6.37DD72 pKa = 6.25YY73 pKa = 11.69DD74 pKa = 5.83DD75 pKa = 6.79DD76 pKa = 6.0YY77 pKa = 12.01DD78 pKa = 6.1DD79 pKa = 6.01YY80 pKa = 12.07DD81 pKa = 4.24DD82 pKa = 4.2EE83 pKa = 5.9PYY85 pKa = 10.61VEE87 pKa = 5.43KK88 pKa = 10.0NTKK91 pKa = 8.72VKK93 pKa = 10.69KK94 pKa = 10.47KK95 pKa = 9.08LTSNQKK101 pKa = 8.83VNRR104 pKa = 11.84AIAILSGVIFCFTAFYY120 pKa = 10.38GAIAISSFAGKK131 pKa = 8.07TKK133 pKa = 10.38AAQEE137 pKa = 4.15APADD141 pKa = 3.83NQTEE145 pKa = 4.17AEE147 pKa = 4.11EE148 pKa = 4.87GFFVSQNSDD157 pKa = 2.59ATISQEE163 pKa = 5.02IKK165 pKa = 9.91DD166 pKa = 4.19TLNSATLEE174 pKa = 4.16PLTTGYY180 pKa = 10.42EE181 pKa = 4.22AIDD184 pKa = 3.44NSVANMLKK192 pKa = 8.65VTCKK196 pKa = 10.91DD197 pKa = 4.25DD198 pKa = 3.07MTTYY202 pKa = 11.17DD203 pKa = 3.82KK204 pKa = 11.32VRR206 pKa = 11.84NIYY209 pKa = 10.59DD210 pKa = 3.39YY211 pKa = 11.78MMFTYY216 pKa = 10.21EE217 pKa = 3.86NADD220 pKa = 3.13ASFVDD225 pKa = 4.0EE226 pKa = 4.58DD227 pKa = 4.62SVYY230 pKa = 11.14DD231 pKa = 3.56FCNKK235 pKa = 9.75YY236 pKa = 10.84SYY238 pKa = 11.11TNYY241 pKa = 9.89FDD243 pKa = 4.82MEE245 pKa = 4.1MLYY248 pKa = 10.56RR249 pKa = 11.84ADD251 pKa = 4.38KK252 pKa = 11.23LMASKK257 pKa = 10.85AGDD260 pKa = 3.46SRR262 pKa = 11.84DD263 pKa = 3.71YY264 pKa = 11.57GSTFAIALRR273 pKa = 11.84ALGLDD278 pKa = 3.67AYY280 pKa = 10.54YY281 pKa = 11.02VATSSDD287 pKa = 3.44TNYY290 pKa = 10.75LAGLDD295 pKa = 3.97TIKK298 pKa = 10.86GYY300 pKa = 8.01TVVVIDD306 pKa = 3.81DD307 pKa = 3.81EE308 pKa = 5.79KK309 pKa = 11.63YY310 pKa = 9.9VFDD313 pKa = 4.81VNAEE317 pKa = 4.46DD318 pKa = 4.82IEE320 pKa = 4.73STDD323 pKa = 3.3SEE325 pKa = 4.72AGVVYY330 pKa = 10.72SFFGKK335 pKa = 10.31DD336 pKa = 3.53FLDD339 pKa = 4.2LPEE342 pKa = 4.52QYY344 pKa = 10.03TLEE347 pKa = 4.29DD348 pKa = 3.53AEE350 pKa = 4.38ASIEE354 pKa = 4.0AFGNFGTLGEE364 pKa = 4.28FGISVVATTKK374 pKa = 10.37SGEE377 pKa = 4.17YY378 pKa = 10.07SSGAVSYY385 pKa = 10.94NSSGEE390 pKa = 4.0NLIEE394 pKa = 4.97ADD396 pKa = 4.53TIGIDD401 pKa = 3.46QGEE404 pKa = 4.38TIYY407 pKa = 10.87FKK409 pKa = 10.74GTVSGSEE416 pKa = 4.0TNTWKK421 pKa = 11.02LIADD425 pKa = 4.09VYY427 pKa = 9.96DD428 pKa = 4.04HH429 pKa = 6.45NWNYY433 pKa = 7.98VTEE436 pKa = 4.16ATLYY440 pKa = 11.15NEE442 pKa = 4.21THH444 pKa = 6.52SNRR447 pKa = 11.84TDD449 pKa = 3.66EE450 pKa = 4.13ITYY453 pKa = 10.12APSRR457 pKa = 11.84GGFIVIRR464 pKa = 11.84FIVTDD469 pKa = 3.73EE470 pKa = 3.96NGRR473 pKa = 11.84TCTVKK478 pKa = 10.62FIADD482 pKa = 3.57VAGDD486 pKa = 3.79DD487 pKa = 4.51YY488 pKa = 11.7YY489 pKa = 11.4FEE491 pKa = 5.71PEE493 pKa = 3.84TTTVYY498 pKa = 8.05EE499 pKa = 4.48TTTEE503 pKa = 4.19EE504 pKa = 4.6YY505 pKa = 7.81TTEE508 pKa = 3.85EE509 pKa = 4.12VTTIEE514 pKa = 3.98EE515 pKa = 4.44TTTVEE520 pKa = 3.76EE521 pKa = 4.51TTTVEE526 pKa = 4.02EE527 pKa = 4.45TTSQEE532 pKa = 3.84EE533 pKa = 4.36TTSQEE538 pKa = 3.9EE539 pKa = 4.35SSSQEE544 pKa = 3.72EE545 pKa = 4.25TSSQEE550 pKa = 3.9EE551 pKa = 4.46TSSQTEE557 pKa = 4.49PEE559 pKa = 4.53PQPQPQPP566 pKa = 3.37
Molecular weight: 64.22 kDa
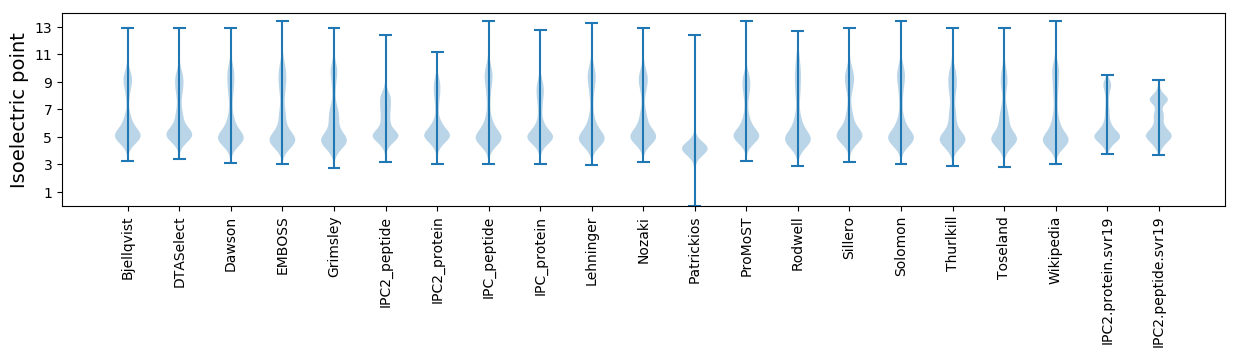
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H9DRP7|A0A1H9DRP7_9FIRM tRNA uridine 5-carboxymethylaminomethyl modification enzyme MnmG OS=Lachnospiraceae bacterium RM5 OX=1200721 GN=mnmG PE=3 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.61KK9 pKa = 7.54RR10 pKa = 11.84QRR12 pKa = 11.84SRR14 pKa = 11.84VHH16 pKa = 6.37GFRR19 pKa = 11.84KK20 pKa = 10.0RR21 pKa = 11.84MSTASGRR28 pKa = 11.84KK29 pKa = 8.42VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.61GRR39 pKa = 11.84KK40 pKa = 8.87KK41 pKa = 10.59LSAA44 pKa = 3.95
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.61KK9 pKa = 7.54RR10 pKa = 11.84QRR12 pKa = 11.84SRR14 pKa = 11.84VHH16 pKa = 6.37GFRR19 pKa = 11.84KK20 pKa = 10.0RR21 pKa = 11.84MSTASGRR28 pKa = 11.84KK29 pKa = 8.42VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.61GRR39 pKa = 11.84KK40 pKa = 8.87KK41 pKa = 10.59LSAA44 pKa = 3.95
Molecular weight: 5.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
697987 |
39 |
4040 |
350.6 |
39.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.783 ± 0.06 | 1.278 ± 0.019 |
6.744 ± 0.053 | 7.762 ± 0.066 |
4.295 ± 0.039 | 6.368 ± 0.058 |
1.358 ± 0.022 | 8.908 ± 0.058 |
8.853 ± 0.066 | 8.126 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.733 ± 0.026 | 6.199 ± 0.054 |
2.575 ± 0.027 | 1.964 ± 0.027 |
3.461 ± 0.04 | 6.237 ± 0.059 |
5.148 ± 0.076 | 6.639 ± 0.044 |
0.71 ± 0.021 | 4.854 ± 0.046 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
