
Peanut chlorotic fan-spot virus
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.22
Get precalculated fractions of proteins
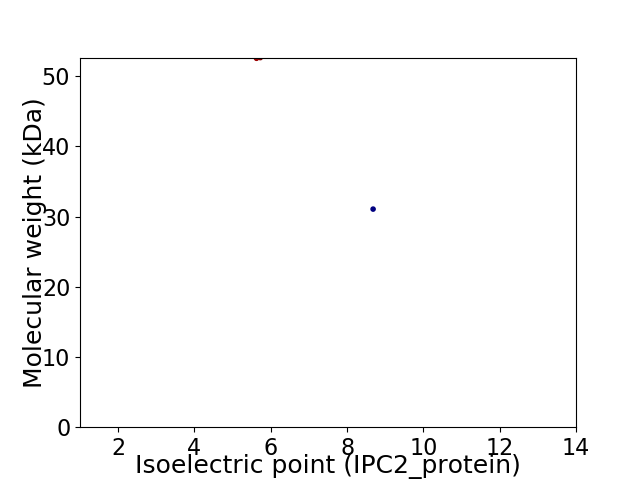
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q9YUD6|Q9YUD6_9VIRU Nonstructural protein NSs OS=Peanut chlorotic fan-spot virus OX=80271 PE=4 SV=1
MM1 pKa = 7.0STEE4 pKa = 3.96SSNPGSTEE12 pKa = 3.61NVALIPEE19 pKa = 4.72PFDD22 pKa = 3.33EE23 pKa = 4.79QYY25 pKa = 11.04CEE27 pKa = 4.02EE28 pKa = 4.58ALTSYY33 pKa = 11.03GNNLDD38 pKa = 3.4KK39 pKa = 11.17SALVDD44 pKa = 3.78VYY46 pKa = 11.9SMFEE50 pKa = 3.81DD51 pKa = 4.07DD52 pKa = 5.31KK53 pKa = 11.8LVFQSIHH60 pKa = 5.14ATNGRR65 pKa = 11.84FKK67 pKa = 9.88ATCAFGHH74 pKa = 6.36SDD76 pKa = 3.28MVLDD80 pKa = 5.13SEE82 pKa = 5.06QEE84 pKa = 4.12LKK86 pKa = 10.35TSISSEE92 pKa = 3.75IFHH95 pKa = 6.4QEE97 pKa = 2.75FDD99 pKa = 4.4AISGINSIISLQGEE113 pKa = 4.5NIHH116 pKa = 6.64VIISHH121 pKa = 6.9PEE123 pKa = 3.8QKK125 pKa = 10.46VKK127 pKa = 10.14SYY129 pKa = 10.82KK130 pKa = 9.61YY131 pKa = 9.85AFHH134 pKa = 6.96GRR136 pKa = 11.84IASNDD141 pKa = 3.26VFPRR145 pKa = 11.84ISGFDD150 pKa = 3.46EE151 pKa = 4.44FRR153 pKa = 11.84SQYY156 pKa = 10.64SCNPYY161 pKa = 10.22KK162 pKa = 10.37FVYY165 pKa = 9.33NPGMLGLEE173 pKa = 4.42SEE175 pKa = 4.8TNTVIFPVEE184 pKa = 3.9KK185 pKa = 10.67LGTLPAHH192 pKa = 7.01ASAFGNNFDD201 pKa = 3.93RR202 pKa = 11.84CHH204 pKa = 6.58FSGSSVPGFMTVKK217 pKa = 10.33AVSEE221 pKa = 4.33SSKK224 pKa = 9.56RR225 pKa = 11.84TQEE228 pKa = 3.77KK229 pKa = 10.03AHH231 pKa = 6.19RR232 pKa = 11.84QLHH235 pKa = 5.33NKK237 pKa = 7.96SLKK240 pKa = 10.39ALEE243 pKa = 4.34IVSNSDD249 pKa = 3.09FRR251 pKa = 11.84STYY254 pKa = 7.99ITKK257 pKa = 8.01TTLKK261 pKa = 10.24ARR263 pKa = 11.84KK264 pKa = 8.7NGMMTIQVQVKK275 pKa = 9.83ISTASLKK282 pKa = 10.32RR283 pKa = 11.84EE284 pKa = 3.95HH285 pKa = 7.04LIAIPDD291 pKa = 4.04DD292 pKa = 3.72PLKK295 pKa = 10.92RR296 pKa = 11.84LVYY299 pKa = 10.41CSSEE303 pKa = 4.17CLGQTSPGLSTFIFEE318 pKa = 5.27VMCLDD323 pKa = 4.02CLVDD327 pKa = 4.43RR328 pKa = 11.84VTPDD332 pKa = 2.92VTYY335 pKa = 10.11FAGALSTCFTDD346 pKa = 4.15EE347 pKa = 4.6PPHH350 pKa = 7.2ILLKK354 pKa = 10.59ARR356 pKa = 11.84SGILSSLADD365 pKa = 3.62SDD367 pKa = 3.49RR368 pKa = 11.84DD369 pKa = 3.7KK370 pKa = 11.29INEE373 pKa = 4.31IICKK377 pKa = 9.95NLVSVHH383 pKa = 6.43LGLATDD389 pKa = 4.77LAKK392 pKa = 10.84KK393 pKa = 10.03LNKK396 pKa = 9.04PINVFTIKK404 pKa = 9.74DD405 pKa = 3.9TSSMTTDD412 pKa = 3.12MVEE415 pKa = 4.04VDD417 pKa = 3.33GKK419 pKa = 10.67KK420 pKa = 10.5YY421 pKa = 10.26RR422 pKa = 11.84VLKK425 pKa = 10.61DD426 pKa = 3.43SNGDD430 pKa = 3.6YY431 pKa = 10.83YY432 pKa = 10.43FTSATFEE439 pKa = 3.99KK440 pKa = 10.13TLIGAYY446 pKa = 9.41KK447 pKa = 9.98SCQTFVDD454 pKa = 3.74YY455 pKa = 11.2CDD457 pKa = 3.63NKK459 pKa = 10.59KK460 pKa = 10.88LSINGDD466 pKa = 3.1NVFIFNN472 pKa = 4.41
MM1 pKa = 7.0STEE4 pKa = 3.96SSNPGSTEE12 pKa = 3.61NVALIPEE19 pKa = 4.72PFDD22 pKa = 3.33EE23 pKa = 4.79QYY25 pKa = 11.04CEE27 pKa = 4.02EE28 pKa = 4.58ALTSYY33 pKa = 11.03GNNLDD38 pKa = 3.4KK39 pKa = 11.17SALVDD44 pKa = 3.78VYY46 pKa = 11.9SMFEE50 pKa = 3.81DD51 pKa = 4.07DD52 pKa = 5.31KK53 pKa = 11.8LVFQSIHH60 pKa = 5.14ATNGRR65 pKa = 11.84FKK67 pKa = 9.88ATCAFGHH74 pKa = 6.36SDD76 pKa = 3.28MVLDD80 pKa = 5.13SEE82 pKa = 5.06QEE84 pKa = 4.12LKK86 pKa = 10.35TSISSEE92 pKa = 3.75IFHH95 pKa = 6.4QEE97 pKa = 2.75FDD99 pKa = 4.4AISGINSIISLQGEE113 pKa = 4.5NIHH116 pKa = 6.64VIISHH121 pKa = 6.9PEE123 pKa = 3.8QKK125 pKa = 10.46VKK127 pKa = 10.14SYY129 pKa = 10.82KK130 pKa = 9.61YY131 pKa = 9.85AFHH134 pKa = 6.96GRR136 pKa = 11.84IASNDD141 pKa = 3.26VFPRR145 pKa = 11.84ISGFDD150 pKa = 3.46EE151 pKa = 4.44FRR153 pKa = 11.84SQYY156 pKa = 10.64SCNPYY161 pKa = 10.22KK162 pKa = 10.37FVYY165 pKa = 9.33NPGMLGLEE173 pKa = 4.42SEE175 pKa = 4.8TNTVIFPVEE184 pKa = 3.9KK185 pKa = 10.67LGTLPAHH192 pKa = 7.01ASAFGNNFDD201 pKa = 3.93RR202 pKa = 11.84CHH204 pKa = 6.58FSGSSVPGFMTVKK217 pKa = 10.33AVSEE221 pKa = 4.33SSKK224 pKa = 9.56RR225 pKa = 11.84TQEE228 pKa = 3.77KK229 pKa = 10.03AHH231 pKa = 6.19RR232 pKa = 11.84QLHH235 pKa = 5.33NKK237 pKa = 7.96SLKK240 pKa = 10.39ALEE243 pKa = 4.34IVSNSDD249 pKa = 3.09FRR251 pKa = 11.84STYY254 pKa = 7.99ITKK257 pKa = 8.01TTLKK261 pKa = 10.24ARR263 pKa = 11.84KK264 pKa = 8.7NGMMTIQVQVKK275 pKa = 9.83ISTASLKK282 pKa = 10.32RR283 pKa = 11.84EE284 pKa = 3.95HH285 pKa = 7.04LIAIPDD291 pKa = 4.04DD292 pKa = 3.72PLKK295 pKa = 10.92RR296 pKa = 11.84LVYY299 pKa = 10.41CSSEE303 pKa = 4.17CLGQTSPGLSTFIFEE318 pKa = 5.27VMCLDD323 pKa = 4.02CLVDD327 pKa = 4.43RR328 pKa = 11.84VTPDD332 pKa = 2.92VTYY335 pKa = 10.11FAGALSTCFTDD346 pKa = 4.15EE347 pKa = 4.6PPHH350 pKa = 7.2ILLKK354 pKa = 10.59ARR356 pKa = 11.84SGILSSLADD365 pKa = 3.62SDD367 pKa = 3.49RR368 pKa = 11.84DD369 pKa = 3.7KK370 pKa = 11.29INEE373 pKa = 4.31IICKK377 pKa = 9.95NLVSVHH383 pKa = 6.43LGLATDD389 pKa = 4.77LAKK392 pKa = 10.84KK393 pKa = 10.03LNKK396 pKa = 9.04PINVFTIKK404 pKa = 9.74DD405 pKa = 3.9TSSMTTDD412 pKa = 3.12MVEE415 pKa = 4.04VDD417 pKa = 3.33GKK419 pKa = 10.67KK420 pKa = 10.5YY421 pKa = 10.26RR422 pKa = 11.84VLKK425 pKa = 10.61DD426 pKa = 3.43SNGDD430 pKa = 3.6YY431 pKa = 10.83YY432 pKa = 10.43FTSATFEE439 pKa = 3.99KK440 pKa = 10.13TLIGAYY446 pKa = 9.41KK447 pKa = 9.98SCQTFVDD454 pKa = 3.74YY455 pKa = 11.2CDD457 pKa = 3.63NKK459 pKa = 10.59KK460 pKa = 10.88LSINGDD466 pKa = 3.1NVFIFNN472 pKa = 4.41
Molecular weight: 52.54 kDa
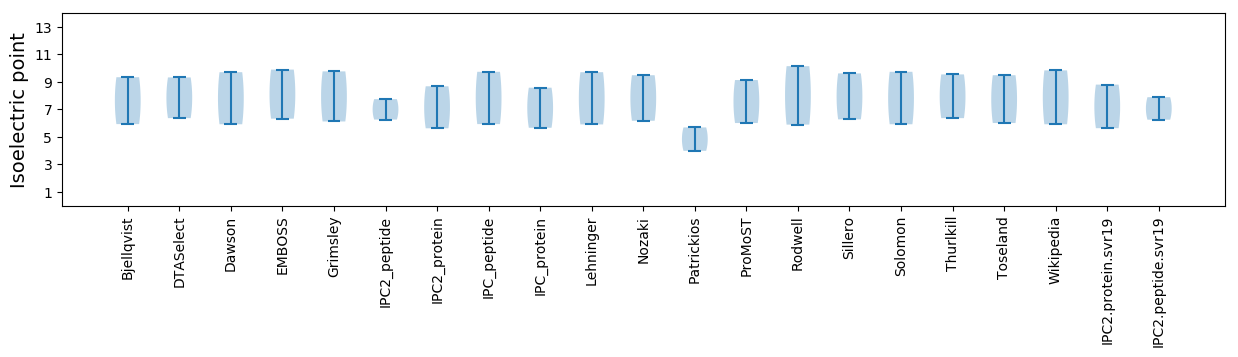
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q9YUD6|Q9YUD6_9VIRU Nonstructural protein NSs OS=Peanut chlorotic fan-spot virus OX=80271 PE=4 SV=1
MM1 pKa = 7.6SKK3 pKa = 9.31TKK5 pKa = 10.66VKK7 pKa = 10.5NDD9 pKa = 3.16KK10 pKa = 10.52EE11 pKa = 4.27LFEE14 pKa = 4.37SLSRR18 pKa = 11.84NAKK21 pKa = 9.74IEE23 pKa = 3.85LSQEE27 pKa = 3.76QANFTFKK34 pKa = 10.89KK35 pKa = 10.26FFSEE39 pKa = 4.04NNSKK43 pKa = 10.55VEE45 pKa = 3.79ISDD48 pKa = 3.31EE49 pKa = 4.19NMILFINNSNKK60 pKa = 8.95MKK62 pKa = 10.7AIGRR66 pKa = 11.84EE67 pKa = 4.15SSVSKK72 pKa = 10.79YY73 pKa = 10.2LGHH76 pKa = 6.76SIVKK80 pKa = 10.31SSPGVNEE87 pKa = 4.21FTWSRR92 pKa = 11.84MDD94 pKa = 3.2SLIRR98 pKa = 11.84MKK100 pKa = 10.78YY101 pKa = 8.12IEE103 pKa = 4.23RR104 pKa = 11.84VKK106 pKa = 10.86DD107 pKa = 3.55YY108 pKa = 11.55GDD110 pKa = 4.13DD111 pKa = 3.49RR112 pKa = 11.84MKK114 pKa = 11.2AEE116 pKa = 4.09NAKK119 pKa = 8.65LTNWLLEE126 pKa = 4.06VFNLHH131 pKa = 6.06TDD133 pKa = 3.11AMSPRR138 pKa = 11.84DD139 pKa = 3.51EE140 pKa = 5.1VIFKK144 pKa = 10.75VVTGGNLNHH153 pKa = 6.95LMGFKK158 pKa = 8.22TTFAHH163 pKa = 7.16AFAIANYY170 pKa = 5.88QHH172 pKa = 6.58KK173 pKa = 10.19RR174 pKa = 11.84SEE176 pKa = 3.93EE177 pKa = 3.78LGIKK181 pKa = 10.49NFDD184 pKa = 3.3TKK186 pKa = 11.2AQLDD190 pKa = 3.73RR191 pKa = 11.84MVVIGEE197 pKa = 4.24RR198 pKa = 11.84CSLLPSPTPKK208 pKa = 10.03TLIEE212 pKa = 4.42SYY214 pKa = 10.43FRR216 pKa = 11.84NAVPKK221 pKa = 10.47AKK223 pKa = 10.03SDD225 pKa = 4.1SKK227 pKa = 11.27DD228 pKa = 3.22QSSKK232 pKa = 11.55YY233 pKa = 10.59NDD235 pKa = 3.72LQNAIAAVNFEE246 pKa = 4.43VPWRR250 pKa = 11.84QVMLYY255 pKa = 9.78QVLNIYY261 pKa = 10.38LLIVKK266 pKa = 10.18AFLFF270 pKa = 4.32
MM1 pKa = 7.6SKK3 pKa = 9.31TKK5 pKa = 10.66VKK7 pKa = 10.5NDD9 pKa = 3.16KK10 pKa = 10.52EE11 pKa = 4.27LFEE14 pKa = 4.37SLSRR18 pKa = 11.84NAKK21 pKa = 9.74IEE23 pKa = 3.85LSQEE27 pKa = 3.76QANFTFKK34 pKa = 10.89KK35 pKa = 10.26FFSEE39 pKa = 4.04NNSKK43 pKa = 10.55VEE45 pKa = 3.79ISDD48 pKa = 3.31EE49 pKa = 4.19NMILFINNSNKK60 pKa = 8.95MKK62 pKa = 10.7AIGRR66 pKa = 11.84EE67 pKa = 4.15SSVSKK72 pKa = 10.79YY73 pKa = 10.2LGHH76 pKa = 6.76SIVKK80 pKa = 10.31SSPGVNEE87 pKa = 4.21FTWSRR92 pKa = 11.84MDD94 pKa = 3.2SLIRR98 pKa = 11.84MKK100 pKa = 10.78YY101 pKa = 8.12IEE103 pKa = 4.23RR104 pKa = 11.84VKK106 pKa = 10.86DD107 pKa = 3.55YY108 pKa = 11.55GDD110 pKa = 4.13DD111 pKa = 3.49RR112 pKa = 11.84MKK114 pKa = 11.2AEE116 pKa = 4.09NAKK119 pKa = 8.65LTNWLLEE126 pKa = 4.06VFNLHH131 pKa = 6.06TDD133 pKa = 3.11AMSPRR138 pKa = 11.84DD139 pKa = 3.51EE140 pKa = 5.1VIFKK144 pKa = 10.75VVTGGNLNHH153 pKa = 6.95LMGFKK158 pKa = 8.22TTFAHH163 pKa = 7.16AFAIANYY170 pKa = 5.88QHH172 pKa = 6.58KK173 pKa = 10.19RR174 pKa = 11.84SEE176 pKa = 3.93EE177 pKa = 3.78LGIKK181 pKa = 10.49NFDD184 pKa = 3.3TKK186 pKa = 11.2AQLDD190 pKa = 3.73RR191 pKa = 11.84MVVIGEE197 pKa = 4.24RR198 pKa = 11.84CSLLPSPTPKK208 pKa = 10.03TLIEE212 pKa = 4.42SYY214 pKa = 10.43FRR216 pKa = 11.84NAVPKK221 pKa = 10.47AKK223 pKa = 10.03SDD225 pKa = 4.1SKK227 pKa = 11.27DD228 pKa = 3.22QSSKK232 pKa = 11.55YY233 pKa = 10.59NDD235 pKa = 3.72LQNAIAAVNFEE246 pKa = 4.43VPWRR250 pKa = 11.84QVMLYY255 pKa = 9.78QVLNIYY261 pKa = 10.38LLIVKK266 pKa = 10.18AFLFF270 pKa = 4.32
Molecular weight: 31.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
742 |
270 |
472 |
371.0 |
41.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.795 ± 0.298 | 1.752 ± 0.822 |
5.795 ± 0.584 | 6.065 ± 0.358 |
6.065 ± 0.138 | 4.447 ± 0.663 |
2.426 ± 0.342 | 6.334 ± 0.243 |
8.221 ± 1.059 | 8.086 ± 0.257 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.695 ± 0.6 | 6.334 ± 1.08 |
3.235 ± 0.382 | 2.83 ± 0.079 |
3.639 ± 0.48 | 10.377 ± 0.666 |
5.795 ± 1.024 | 6.469 ± 0.118 |
0.404 ± 0.421 | 3.235 ± 0.162 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
