
Capybara microvirus Cap3_SP_641
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
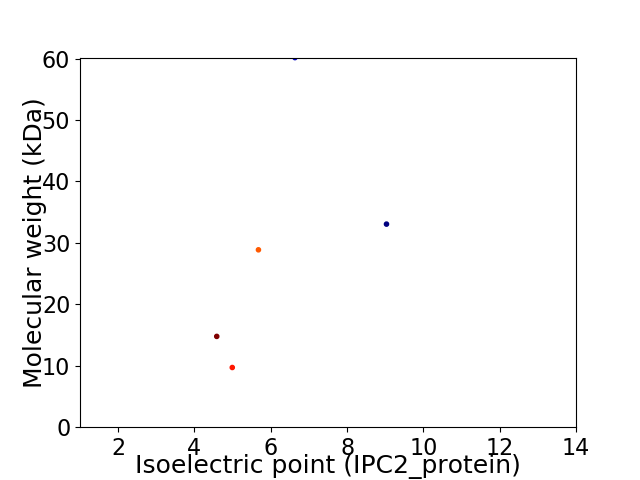
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
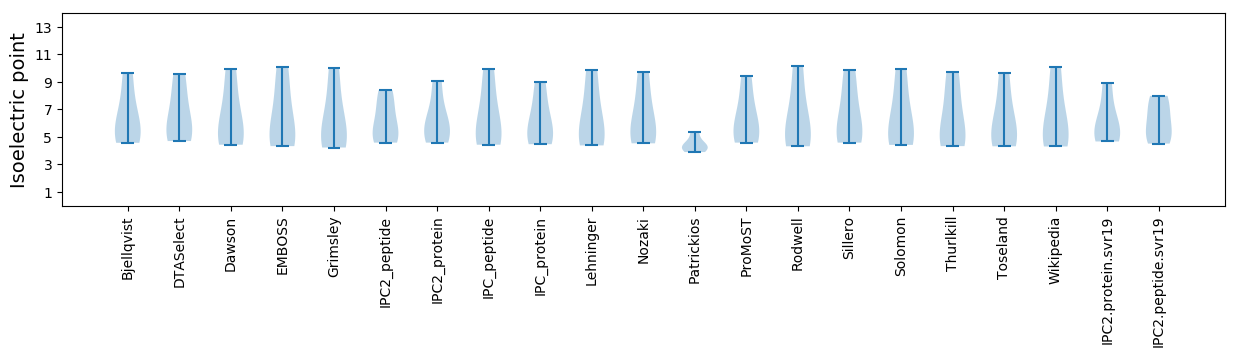
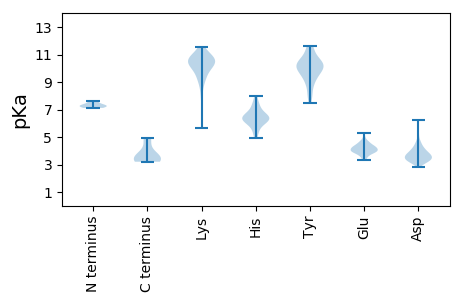
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W8Z7|A0A4P8W8Z7_9VIRU Minor capsid protein OS=Capybara microvirus Cap3_SP_641 OX=2585485 PE=4 SV=1
MM1 pKa = 7.26NAVSDD6 pKa = 4.04EE7 pKa = 4.23TALRR11 pKa = 11.84CNDD14 pKa = 3.55PSLTQQQFKK23 pKa = 10.91EE24 pKa = 4.06EE25 pKa = 3.99SDD27 pKa = 3.35INNIVDD33 pKa = 4.48LFMKK37 pKa = 9.77TGHH40 pKa = 6.95LPTPVSMPQYY50 pKa = 10.25VDD52 pKa = 3.3YY53 pKa = 11.32EE54 pKa = 4.79GVFDD58 pKa = 5.12FQSAMNAVRR67 pKa = 11.84QADD70 pKa = 4.14EE71 pKa = 3.91NFMRR75 pKa = 11.84MDD77 pKa = 3.41AKK79 pKa = 10.73LRR81 pKa = 11.84ARR83 pKa = 11.84FHH85 pKa = 6.67NSPQEE90 pKa = 3.9FLEE93 pKa = 5.28FFADD97 pKa = 3.76PNNTEE102 pKa = 3.67EE103 pKa = 4.31AVRR106 pKa = 11.84LGLAIAKK113 pKa = 9.16PKK115 pKa = 10.5EE116 pKa = 3.94FTSPVVEE123 pKa = 4.2SAKK126 pKa = 10.83ADD128 pKa = 3.59GAASS132 pKa = 3.37
MM1 pKa = 7.26NAVSDD6 pKa = 4.04EE7 pKa = 4.23TALRR11 pKa = 11.84CNDD14 pKa = 3.55PSLTQQQFKK23 pKa = 10.91EE24 pKa = 4.06EE25 pKa = 3.99SDD27 pKa = 3.35INNIVDD33 pKa = 4.48LFMKK37 pKa = 9.77TGHH40 pKa = 6.95LPTPVSMPQYY50 pKa = 10.25VDD52 pKa = 3.3YY53 pKa = 11.32EE54 pKa = 4.79GVFDD58 pKa = 5.12FQSAMNAVRR67 pKa = 11.84QADD70 pKa = 4.14EE71 pKa = 3.91NFMRR75 pKa = 11.84MDD77 pKa = 3.41AKK79 pKa = 10.73LRR81 pKa = 11.84ARR83 pKa = 11.84FHH85 pKa = 6.67NSPQEE90 pKa = 3.9FLEE93 pKa = 5.28FFADD97 pKa = 3.76PNNTEE102 pKa = 3.67EE103 pKa = 4.31AVRR106 pKa = 11.84LGLAIAKK113 pKa = 9.16PKK115 pKa = 10.5EE116 pKa = 3.94FTSPVVEE123 pKa = 4.2SAKK126 pKa = 10.83ADD128 pKa = 3.59GAASS132 pKa = 3.37
Molecular weight: 14.78 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W5D6|A0A4P8W5D6_9VIRU Major capsid protein OS=Capybara microvirus Cap3_SP_641 OX=2585485 PE=3 SV=1
MM1 pKa = 7.25CTALWQHH8 pKa = 6.85PSHH11 pKa = 6.76GPTKK15 pKa = 10.39CGQCVEE21 pKa = 4.27CRR23 pKa = 11.84LAYY26 pKa = 9.97SRR28 pKa = 11.84EE29 pKa = 3.38WAIRR33 pKa = 11.84ITHH36 pKa = 5.98EE37 pKa = 4.0QQMHH41 pKa = 6.22KK42 pKa = 10.07VSCMLNLTYY51 pKa = 10.74DD52 pKa = 3.92DD53 pKa = 3.65EE54 pKa = 4.95HH55 pKa = 8.01LPKK58 pKa = 10.38HH59 pKa = 5.35GQLHH63 pKa = 6.26KK64 pKa = 10.88EE65 pKa = 3.82HH66 pKa = 6.3LQKK69 pKa = 10.66FFKK72 pKa = 10.52RR73 pKa = 11.84MRR75 pKa = 11.84KK76 pKa = 9.02AGYY79 pKa = 8.83KK80 pKa = 9.44FRR82 pKa = 11.84YY83 pKa = 7.94VASGEE88 pKa = 4.16YY89 pKa = 10.49GDD91 pKa = 4.08VSRR94 pKa = 11.84RR95 pKa = 11.84PHH97 pKa = 4.95FHH99 pKa = 6.29IALFGVDD106 pKa = 4.27FDD108 pKa = 4.72ADD110 pKa = 3.61RR111 pKa = 11.84VPFGRR116 pKa = 11.84SGGDD120 pKa = 2.79RR121 pKa = 11.84TYY123 pKa = 9.44TSSRR127 pKa = 11.84VLHH130 pKa = 6.3LWPFGNHH137 pKa = 7.51LIGTLNFEE145 pKa = 4.02SAAYY149 pKa = 8.06IARR152 pKa = 11.84YY153 pKa = 8.94IMKK156 pKa = 10.13KK157 pKa = 9.76IKK159 pKa = 10.26GPNASPVPLFNDD171 pKa = 3.73DD172 pKa = 3.37EE173 pKa = 4.57TGEE176 pKa = 4.28VVMPNPEE183 pKa = 3.95FLLVSKK189 pKa = 10.83GRR191 pKa = 11.84GKK193 pKa = 10.75GQGIGGSWFRR203 pKa = 11.84DD204 pKa = 3.39YY205 pKa = 11.64FMTDD209 pKa = 3.26VFPHH213 pKa = 6.57AAVITPQRR221 pKa = 11.84SKK223 pKa = 11.5APVPRR228 pKa = 11.84YY229 pKa = 10.04YY230 pKa = 9.65KK231 pKa = 10.24TLLKK235 pKa = 10.7EE236 pKa = 4.16LGSDD240 pKa = 4.49LALDD244 pKa = 3.55MQFRR248 pKa = 11.84SSVRR252 pKa = 11.84ADD254 pKa = 3.29MEE256 pKa = 4.19LEE258 pKa = 3.66RR259 pKa = 11.84RR260 pKa = 11.84MLEE263 pKa = 4.02DD264 pKa = 3.19QPVRR268 pKa = 11.84KK269 pKa = 9.02IARR272 pKa = 11.84QIVSQSRR279 pKa = 11.84NNLSKK284 pKa = 10.93RR285 pKa = 11.84ILL287 pKa = 3.61
MM1 pKa = 7.25CTALWQHH8 pKa = 6.85PSHH11 pKa = 6.76GPTKK15 pKa = 10.39CGQCVEE21 pKa = 4.27CRR23 pKa = 11.84LAYY26 pKa = 9.97SRR28 pKa = 11.84EE29 pKa = 3.38WAIRR33 pKa = 11.84ITHH36 pKa = 5.98EE37 pKa = 4.0QQMHH41 pKa = 6.22KK42 pKa = 10.07VSCMLNLTYY51 pKa = 10.74DD52 pKa = 3.92DD53 pKa = 3.65EE54 pKa = 4.95HH55 pKa = 8.01LPKK58 pKa = 10.38HH59 pKa = 5.35GQLHH63 pKa = 6.26KK64 pKa = 10.88EE65 pKa = 3.82HH66 pKa = 6.3LQKK69 pKa = 10.66FFKK72 pKa = 10.52RR73 pKa = 11.84MRR75 pKa = 11.84KK76 pKa = 9.02AGYY79 pKa = 8.83KK80 pKa = 9.44FRR82 pKa = 11.84YY83 pKa = 7.94VASGEE88 pKa = 4.16YY89 pKa = 10.49GDD91 pKa = 4.08VSRR94 pKa = 11.84RR95 pKa = 11.84PHH97 pKa = 4.95FHH99 pKa = 6.29IALFGVDD106 pKa = 4.27FDD108 pKa = 4.72ADD110 pKa = 3.61RR111 pKa = 11.84VPFGRR116 pKa = 11.84SGGDD120 pKa = 2.79RR121 pKa = 11.84TYY123 pKa = 9.44TSSRR127 pKa = 11.84VLHH130 pKa = 6.3LWPFGNHH137 pKa = 7.51LIGTLNFEE145 pKa = 4.02SAAYY149 pKa = 8.06IARR152 pKa = 11.84YY153 pKa = 8.94IMKK156 pKa = 10.13KK157 pKa = 9.76IKK159 pKa = 10.26GPNASPVPLFNDD171 pKa = 3.73DD172 pKa = 3.37EE173 pKa = 4.57TGEE176 pKa = 4.28VVMPNPEE183 pKa = 3.95FLLVSKK189 pKa = 10.83GRR191 pKa = 11.84GKK193 pKa = 10.75GQGIGGSWFRR203 pKa = 11.84DD204 pKa = 3.39YY205 pKa = 11.64FMTDD209 pKa = 3.26VFPHH213 pKa = 6.57AAVITPQRR221 pKa = 11.84SKK223 pKa = 11.5APVPRR228 pKa = 11.84YY229 pKa = 10.04YY230 pKa = 9.65KK231 pKa = 10.24TLLKK235 pKa = 10.7EE236 pKa = 4.16LGSDD240 pKa = 4.49LALDD244 pKa = 3.55MQFRR248 pKa = 11.84SSVRR252 pKa = 11.84ADD254 pKa = 3.29MEE256 pKa = 4.19LEE258 pKa = 3.66RR259 pKa = 11.84RR260 pKa = 11.84MLEE263 pKa = 4.02DD264 pKa = 3.19QPVRR268 pKa = 11.84KK269 pKa = 9.02IARR272 pKa = 11.84QIVSQSRR279 pKa = 11.84NNLSKK284 pKa = 10.93RR285 pKa = 11.84ILL287 pKa = 3.61
Molecular weight: 33.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1330 |
85 |
552 |
266.0 |
29.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.624 ± 1.97 | 0.602 ± 0.337 |
5.564 ± 0.621 | 3.835 ± 0.925 |
5.263 ± 0.793 | 7.594 ± 1.025 |
2.03 ± 0.779 | 4.06 ± 0.353 |
3.985 ± 0.897 | 8.346 ± 0.524 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.632 ± 0.512 | 5.94 ± 0.871 |
5.038 ± 0.699 | 5.714 ± 0.877 |
5.414 ± 1.004 | 7.82 ± 0.687 |
5.338 ± 0.809 | 6.692 ± 1.05 |
0.977 ± 0.284 | 3.534 ± 0.459 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
