
Limnohabitans sp. 103DPR2
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Comamonadaceae; Limnohabitans; unclassified Limnohabitans
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins
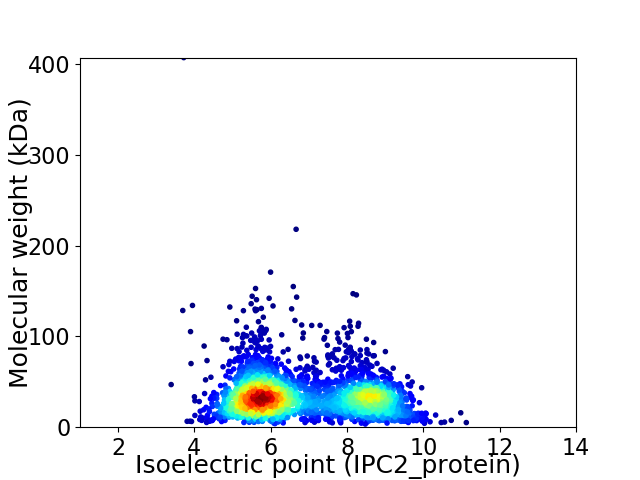
Virtual 2D-PAGE plot for 2804 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0P0MHY7|A0A0P0MHY7_9BURK HemY_N domain-containing protein OS=Limnohabitans sp. 103DPR2 OX=1678129 GN=L103DPR2_02444 PE=4 SV=1
MM1 pKa = 7.53PIKK4 pKa = 10.79YY5 pKa = 7.64GTNGNDD11 pKa = 3.02NPLRR15 pKa = 11.84GTSGNDD21 pKa = 3.04SLYY24 pKa = 11.36GLAGDD29 pKa = 5.39DD30 pKa = 5.35FILTEE35 pKa = 4.61DD36 pKa = 3.84GEE38 pKa = 5.11DD39 pKa = 3.62YY40 pKa = 11.54VEE42 pKa = 5.77AGDD45 pKa = 5.92GDD47 pKa = 4.77DD48 pKa = 4.64EE49 pKa = 5.16VNGYY53 pKa = 10.78DD54 pKa = 4.14GVGGSYY60 pKa = 9.69TYY62 pKa = 11.24YY63 pKa = 9.86PVAGIKK69 pKa = 9.01TIHH72 pKa = 6.52GGNGNDD78 pKa = 3.51FLVGGSAGDD87 pKa = 3.48VLYY90 pKa = 10.99GDD92 pKa = 4.61EE93 pKa = 6.01GNDD96 pKa = 3.26QLYY99 pKa = 10.81GRR101 pKa = 11.84GGNDD105 pKa = 2.9ILSGGPGADD114 pKa = 3.75YY115 pKa = 11.39LNGGPGDD122 pKa = 3.62DD123 pKa = 3.74TYY125 pKa = 11.67YY126 pKa = 11.11VSDD129 pKa = 3.13IHH131 pKa = 8.09DD132 pKa = 4.2VIEE135 pKa = 4.8DD136 pKa = 3.49VSGTDD141 pKa = 3.06TAYY144 pKa = 10.84VATSFVKK151 pKa = 10.35IPSSIEE157 pKa = 3.47KK158 pKa = 10.46VIYY161 pKa = 9.98TDD163 pKa = 4.38GAQSLPYY170 pKa = 9.53WVDD173 pKa = 2.85ALLPDD178 pKa = 4.19EE179 pKa = 4.64AAGNAFEE186 pKa = 4.79SLLGSAHH193 pKa = 6.25TYY195 pKa = 10.19FYY197 pKa = 10.36TFPTSLPTYY206 pKa = 7.41DD207 pKa = 3.36TNYY210 pKa = 10.23SHH212 pKa = 7.23GLGFKK217 pKa = 10.33PFTSTQMARR226 pKa = 11.84AEE228 pKa = 3.99AALSIVSSVIDD239 pKa = 3.08VHH241 pKa = 6.31FQKK244 pKa = 9.98TNNPGVLNTFVFANNDD260 pKa = 3.51QPSSAGSGNFPSDD273 pKa = 3.5YY274 pKa = 10.34MIGSDD279 pKa = 4.78LYY281 pKa = 11.29FDD283 pKa = 4.14NSSLNAAFADD293 pKa = 4.05RR294 pKa = 11.84TYY296 pKa = 11.55GALTLIHH303 pKa = 6.93EE304 pKa = 4.83IGHH307 pKa = 6.01GLGLEE312 pKa = 4.45HH313 pKa = 7.29PFSHH317 pKa = 6.42AQAGSSSVSDD327 pKa = 3.7PPYY330 pKa = 9.85LTGTEE335 pKa = 4.59EE336 pKa = 4.0STAWTVMSYY345 pKa = 11.42NDD347 pKa = 4.81APAQYY352 pKa = 9.53YY353 pKa = 10.82LSFSPLDD360 pKa = 3.52IAALQYY366 pKa = 10.56IYY368 pKa = 10.89GPSKK372 pKa = 9.91TSRR375 pKa = 11.84TGNDD379 pKa = 3.2TYY381 pKa = 11.05KK382 pKa = 10.9VSATEE387 pKa = 5.0PNFIWDD393 pKa = 3.64GAGVDD398 pKa = 4.03TLDD401 pKa = 5.21ASNLNQGSTLYY412 pKa = 8.75LTPGYY417 pKa = 8.95WGYY420 pKa = 11.43VGNNKK425 pKa = 8.3ATNITAAGQVTVNFGSAIEE444 pKa = 4.15NLTGSSFADD453 pKa = 2.99KK454 pKa = 10.9LYY456 pKa = 11.12GNEE459 pKa = 5.2LGNQMSGGMGNDD471 pKa = 3.32WLEE474 pKa = 3.86GWAGDD479 pKa = 3.74DD480 pKa = 3.94TLVGGQGDD488 pKa = 4.04DD489 pKa = 3.55QLQGGSGIDD498 pKa = 3.18TALFGGAYY506 pKa = 10.12ASYY509 pKa = 9.23TFEE512 pKa = 4.28NTSSTFSVKK521 pKa = 10.3DD522 pKa = 3.15KK523 pKa = 10.65RR524 pKa = 11.84ANADD528 pKa = 3.75GIDD531 pKa = 3.45VLTSVEE537 pKa = 4.03RR538 pKa = 11.84LKK540 pKa = 11.3FSDD543 pKa = 3.27KK544 pKa = 10.94SVAIDD549 pKa = 3.61LDD551 pKa = 4.21GNAGIVVKK559 pKa = 10.8VIGAVLGSDD568 pKa = 3.34AVKK571 pKa = 9.89TPGIVGTGLRR581 pKa = 11.84YY582 pKa = 9.9VDD584 pKa = 4.25NGMSYY589 pKa = 11.7ADD591 pKa = 4.58LGLTALNAVGAMTPDD606 pKa = 4.85AIVSTLWRR614 pKa = 11.84NVVGSIASATEE625 pKa = 3.47KK626 pKa = 10.94APYY629 pKa = 10.62LKK631 pKa = 10.03MLADD635 pKa = 3.67GTKK638 pKa = 10.33PGDD641 pKa = 3.76LVVLAGDD648 pKa = 4.65FSLNMNKK655 pKa = 9.63IGLMGLAQTGIEE667 pKa = 4.32FSS669 pKa = 3.51
MM1 pKa = 7.53PIKK4 pKa = 10.79YY5 pKa = 7.64GTNGNDD11 pKa = 3.02NPLRR15 pKa = 11.84GTSGNDD21 pKa = 3.04SLYY24 pKa = 11.36GLAGDD29 pKa = 5.39DD30 pKa = 5.35FILTEE35 pKa = 4.61DD36 pKa = 3.84GEE38 pKa = 5.11DD39 pKa = 3.62YY40 pKa = 11.54VEE42 pKa = 5.77AGDD45 pKa = 5.92GDD47 pKa = 4.77DD48 pKa = 4.64EE49 pKa = 5.16VNGYY53 pKa = 10.78DD54 pKa = 4.14GVGGSYY60 pKa = 9.69TYY62 pKa = 11.24YY63 pKa = 9.86PVAGIKK69 pKa = 9.01TIHH72 pKa = 6.52GGNGNDD78 pKa = 3.51FLVGGSAGDD87 pKa = 3.48VLYY90 pKa = 10.99GDD92 pKa = 4.61EE93 pKa = 6.01GNDD96 pKa = 3.26QLYY99 pKa = 10.81GRR101 pKa = 11.84GGNDD105 pKa = 2.9ILSGGPGADD114 pKa = 3.75YY115 pKa = 11.39LNGGPGDD122 pKa = 3.62DD123 pKa = 3.74TYY125 pKa = 11.67YY126 pKa = 11.11VSDD129 pKa = 3.13IHH131 pKa = 8.09DD132 pKa = 4.2VIEE135 pKa = 4.8DD136 pKa = 3.49VSGTDD141 pKa = 3.06TAYY144 pKa = 10.84VATSFVKK151 pKa = 10.35IPSSIEE157 pKa = 3.47KK158 pKa = 10.46VIYY161 pKa = 9.98TDD163 pKa = 4.38GAQSLPYY170 pKa = 9.53WVDD173 pKa = 2.85ALLPDD178 pKa = 4.19EE179 pKa = 4.64AAGNAFEE186 pKa = 4.79SLLGSAHH193 pKa = 6.25TYY195 pKa = 10.19FYY197 pKa = 10.36TFPTSLPTYY206 pKa = 7.41DD207 pKa = 3.36TNYY210 pKa = 10.23SHH212 pKa = 7.23GLGFKK217 pKa = 10.33PFTSTQMARR226 pKa = 11.84AEE228 pKa = 3.99AALSIVSSVIDD239 pKa = 3.08VHH241 pKa = 6.31FQKK244 pKa = 9.98TNNPGVLNTFVFANNDD260 pKa = 3.51QPSSAGSGNFPSDD273 pKa = 3.5YY274 pKa = 10.34MIGSDD279 pKa = 4.78LYY281 pKa = 11.29FDD283 pKa = 4.14NSSLNAAFADD293 pKa = 4.05RR294 pKa = 11.84TYY296 pKa = 11.55GALTLIHH303 pKa = 6.93EE304 pKa = 4.83IGHH307 pKa = 6.01GLGLEE312 pKa = 4.45HH313 pKa = 7.29PFSHH317 pKa = 6.42AQAGSSSVSDD327 pKa = 3.7PPYY330 pKa = 9.85LTGTEE335 pKa = 4.59EE336 pKa = 4.0STAWTVMSYY345 pKa = 11.42NDD347 pKa = 4.81APAQYY352 pKa = 9.53YY353 pKa = 10.82LSFSPLDD360 pKa = 3.52IAALQYY366 pKa = 10.56IYY368 pKa = 10.89GPSKK372 pKa = 9.91TSRR375 pKa = 11.84TGNDD379 pKa = 3.2TYY381 pKa = 11.05KK382 pKa = 10.9VSATEE387 pKa = 5.0PNFIWDD393 pKa = 3.64GAGVDD398 pKa = 4.03TLDD401 pKa = 5.21ASNLNQGSTLYY412 pKa = 8.75LTPGYY417 pKa = 8.95WGYY420 pKa = 11.43VGNNKK425 pKa = 8.3ATNITAAGQVTVNFGSAIEE444 pKa = 4.15NLTGSSFADD453 pKa = 2.99KK454 pKa = 10.9LYY456 pKa = 11.12GNEE459 pKa = 5.2LGNQMSGGMGNDD471 pKa = 3.32WLEE474 pKa = 3.86GWAGDD479 pKa = 3.74DD480 pKa = 3.94TLVGGQGDD488 pKa = 4.04DD489 pKa = 3.55QLQGGSGIDD498 pKa = 3.18TALFGGAYY506 pKa = 10.12ASYY509 pKa = 9.23TFEE512 pKa = 4.28NTSSTFSVKK521 pKa = 10.3DD522 pKa = 3.15KK523 pKa = 10.65RR524 pKa = 11.84ANADD528 pKa = 3.75GIDD531 pKa = 3.45VLTSVEE537 pKa = 4.03RR538 pKa = 11.84LKK540 pKa = 11.3FSDD543 pKa = 3.27KK544 pKa = 10.94SVAIDD549 pKa = 3.61LDD551 pKa = 4.21GNAGIVVKK559 pKa = 10.8VIGAVLGSDD568 pKa = 3.34AVKK571 pKa = 9.89TPGIVGTGLRR581 pKa = 11.84YY582 pKa = 9.9VDD584 pKa = 4.25NGMSYY589 pKa = 11.7ADD591 pKa = 4.58LGLTALNAVGAMTPDD606 pKa = 4.85AIVSTLWRR614 pKa = 11.84NVVGSIASATEE625 pKa = 3.47KK626 pKa = 10.94APYY629 pKa = 10.62LKK631 pKa = 10.03MLADD635 pKa = 3.67GTKK638 pKa = 10.33PGDD641 pKa = 3.76LVVLAGDD648 pKa = 4.65FSLNMNKK655 pKa = 9.63IGLMGLAQTGIEE667 pKa = 4.32FSS669 pKa = 3.51
Molecular weight: 70.06 kDa
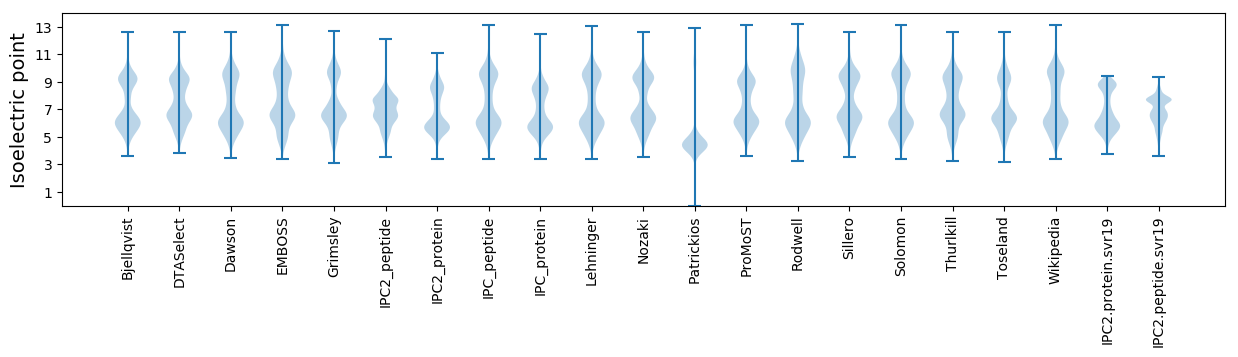
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0P0MCP3|A0A0P0MCP3_9BURK Major Facilitator Superfamily protein OS=Limnohabitans sp. 103DPR2 OX=1678129 GN=L103DPR2_00246 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.25QPSKK9 pKa = 7.79TRR11 pKa = 11.84RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.79GFLVRR21 pKa = 11.84MKK23 pKa = 9.7TRR25 pKa = 11.84GGRR28 pKa = 11.84AVINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.74GRR39 pKa = 11.84KK40 pKa = 8.8RR41 pKa = 11.84LAAA44 pKa = 4.42
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.25QPSKK9 pKa = 7.79TRR11 pKa = 11.84RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.79GFLVRR21 pKa = 11.84MKK23 pKa = 9.7TRR25 pKa = 11.84GGRR28 pKa = 11.84AVINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.74GRR39 pKa = 11.84KK40 pKa = 8.8RR41 pKa = 11.84LAAA44 pKa = 4.42
Molecular weight: 5.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
922556 |
31 |
4090 |
329.0 |
35.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.25 ± 0.058 | 0.964 ± 0.015 |
4.99 ± 0.034 | 5.213 ± 0.042 |
3.757 ± 0.028 | 7.89 ± 0.04 |
2.352 ± 0.026 | 4.886 ± 0.033 |
4.696 ± 0.039 | 10.384 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.934 ± 0.027 | 3.347 ± 0.033 |
4.851 ± 0.031 | 4.591 ± 0.036 |
5.161 ± 0.041 | 5.956 ± 0.043 |
5.397 ± 0.056 | 7.547 ± 0.031 |
1.533 ± 0.025 | 2.3 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
