
Bacteroidales bacterium KA00344
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; unclassified Bacteroidales
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins
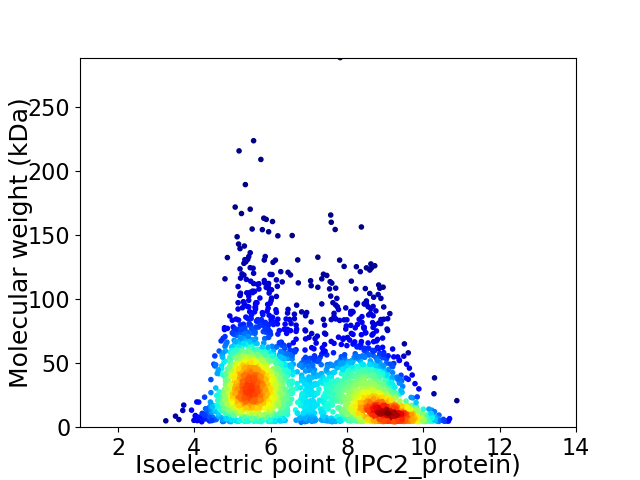
Virtual 2D-PAGE plot for 2869 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
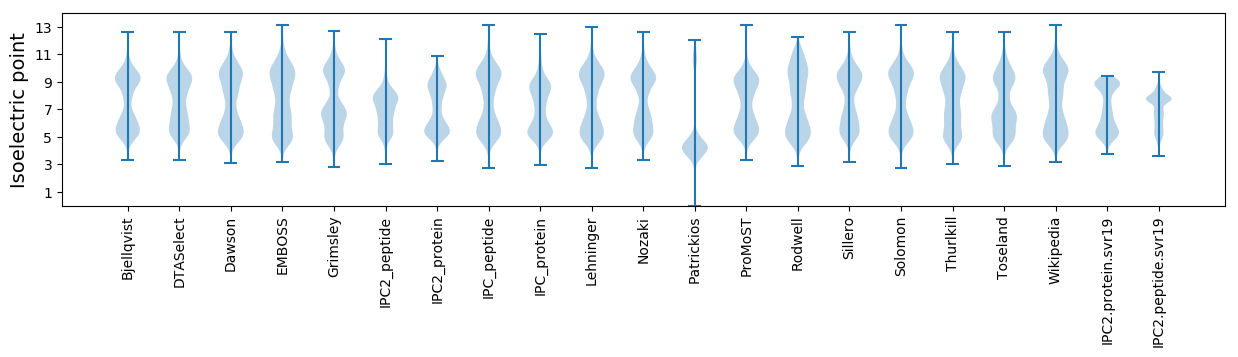
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A133YIG9|A0A133YIG9_9BACT Uncharacterized protein OS=Bacteroidales bacterium KA00344 OX=1497954 GN=HMPREF1870_01375 PE=4 SV=1
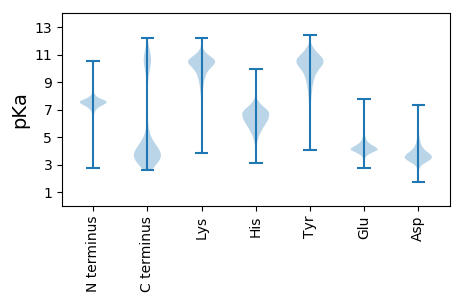
MM1 pKa = 7.65TYY3 pKa = 9.77FCKK6 pKa = 9.59SHH8 pKa = 6.52EE9 pKa = 4.24YY10 pKa = 10.49VRR12 pKa = 11.84VEE14 pKa = 4.41GEE16 pKa = 3.59FGYY19 pKa = 10.32IGITDD24 pKa = 3.95YY25 pKa = 11.29AQKK28 pKa = 11.21ALGNIVYY35 pKa = 10.47VDD37 pKa = 3.88MPDD40 pKa = 3.24VDD42 pKa = 5.32DD43 pKa = 4.74EE44 pKa = 4.68VTAGEE49 pKa = 4.33DD50 pKa = 3.39FGAVEE55 pKa = 4.41SVKK58 pKa = 10.62AASDD62 pKa = 3.47LMSPVSGVVVEE73 pKa = 4.31VNEE76 pKa = 4.1ALEE79 pKa = 4.93DD80 pKa = 3.9EE81 pKa = 5.08PEE83 pKa = 6.02LINQDD88 pKa = 4.53ANANWIIKK96 pKa = 9.93VKK98 pKa = 10.76LSDD101 pKa = 3.77VSEE104 pKa = 4.95LDD106 pKa = 3.7DD107 pKa = 5.87LMDD110 pKa = 4.02EE111 pKa = 4.31EE112 pKa = 6.48AYY114 pKa = 10.48LASVEE119 pKa = 4.12
MM1 pKa = 7.65TYY3 pKa = 9.77FCKK6 pKa = 9.59SHH8 pKa = 6.52EE9 pKa = 4.24YY10 pKa = 10.49VRR12 pKa = 11.84VEE14 pKa = 4.41GEE16 pKa = 3.59FGYY19 pKa = 10.32IGITDD24 pKa = 3.95YY25 pKa = 11.29AQKK28 pKa = 11.21ALGNIVYY35 pKa = 10.47VDD37 pKa = 3.88MPDD40 pKa = 3.24VDD42 pKa = 5.32DD43 pKa = 4.74EE44 pKa = 4.68VTAGEE49 pKa = 4.33DD50 pKa = 3.39FGAVEE55 pKa = 4.41SVKK58 pKa = 10.62AASDD62 pKa = 3.47LMSPVSGVVVEE73 pKa = 4.31VNEE76 pKa = 4.1ALEE79 pKa = 4.93DD80 pKa = 3.9EE81 pKa = 5.08PEE83 pKa = 6.02LINQDD88 pKa = 4.53ANANWIIKK96 pKa = 9.93VKK98 pKa = 10.76LSDD101 pKa = 3.77VSEE104 pKa = 4.95LDD106 pKa = 3.7DD107 pKa = 5.87LMDD110 pKa = 4.02EE111 pKa = 4.31EE112 pKa = 6.48AYY114 pKa = 10.48LASVEE119 pKa = 4.12
Molecular weight: 13.09 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A133Y580|A0A133Y580_9BACT Alanine--tRNA ligase OS=Bacteroidales bacterium KA00344 OX=1497954 GN=alaS PE=3 SV=1
MM1 pKa = 7.65LIVLIEE7 pKa = 3.72AVIRR11 pKa = 11.84KK12 pKa = 9.19RR13 pKa = 11.84ADD15 pKa = 3.1SVLKK19 pKa = 10.79VLAPASKK26 pKa = 10.72VKK28 pKa = 9.9ATTKK32 pKa = 8.74TAHH35 pKa = 6.98HH36 pKa = 6.43ITTIISHH43 pKa = 6.33VATASAKK50 pKa = 10.03AVTVLVTTIRR60 pKa = 11.84RR61 pKa = 11.84AVISLVRR68 pKa = 11.84VAIVPVITTIIKK80 pKa = 9.65RR81 pKa = 11.84AAISLVRR88 pKa = 11.84VVIVPVITTIIKK100 pKa = 9.65RR101 pKa = 11.84AAISLVRR108 pKa = 11.84VVIVPAIITIIRR120 pKa = 11.84RR121 pKa = 11.84ATISLVRR128 pKa = 11.84VVIAPVIITIIRR140 pKa = 11.84RR141 pKa = 11.84VAINLVRR148 pKa = 11.84AVIVPVIITIIRR160 pKa = 11.84RR161 pKa = 11.84AVINRR166 pKa = 11.84VATDD170 pKa = 3.07VRR172 pKa = 11.84VVINRR177 pKa = 11.84AVTVGRR183 pKa = 11.84TMIPMPNITT192 pKa = 4.17
MM1 pKa = 7.65LIVLIEE7 pKa = 3.72AVIRR11 pKa = 11.84KK12 pKa = 9.19RR13 pKa = 11.84ADD15 pKa = 3.1SVLKK19 pKa = 10.79VLAPASKK26 pKa = 10.72VKK28 pKa = 9.9ATTKK32 pKa = 8.74TAHH35 pKa = 6.98HH36 pKa = 6.43ITTIISHH43 pKa = 6.33VATASAKK50 pKa = 10.03AVTVLVTTIRR60 pKa = 11.84RR61 pKa = 11.84AVISLVRR68 pKa = 11.84VAIVPVITTIIKK80 pKa = 9.65RR81 pKa = 11.84AAISLVRR88 pKa = 11.84VVIVPVITTIIKK100 pKa = 9.65RR101 pKa = 11.84AAISLVRR108 pKa = 11.84VVIVPAIITIIRR120 pKa = 11.84RR121 pKa = 11.84ATISLVRR128 pKa = 11.84VVIAPVIITIIRR140 pKa = 11.84RR141 pKa = 11.84VAINLVRR148 pKa = 11.84AVIVPVIITIIRR160 pKa = 11.84RR161 pKa = 11.84AVINRR166 pKa = 11.84VATDD170 pKa = 3.07VRR172 pKa = 11.84VVINRR177 pKa = 11.84AVTVGRR183 pKa = 11.84TMIPMPNITT192 pKa = 4.17
Molecular weight: 20.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
880993 |
35 |
2545 |
307.1 |
34.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.102 ± 0.047 | 1.274 ± 0.019 |
5.746 ± 0.034 | 5.959 ± 0.046 |
4.511 ± 0.028 | 6.764 ± 0.044 |
2.112 ± 0.021 | 6.714 ± 0.045 |
6.682 ± 0.038 | 8.755 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.003 ± 0.024 | 5.246 ± 0.046 |
3.64 ± 0.023 | 3.659 ± 0.025 |
4.967 ± 0.03 | 6.032 ± 0.041 |
5.714 ± 0.037 | 6.527 ± 0.033 |
1.258 ± 0.019 | 4.334 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
