
Corallococcus sp. CAG:1435
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Myxococcales; Cystobacterineae; Myxococcaceae; Corallococcus; environmental samples
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
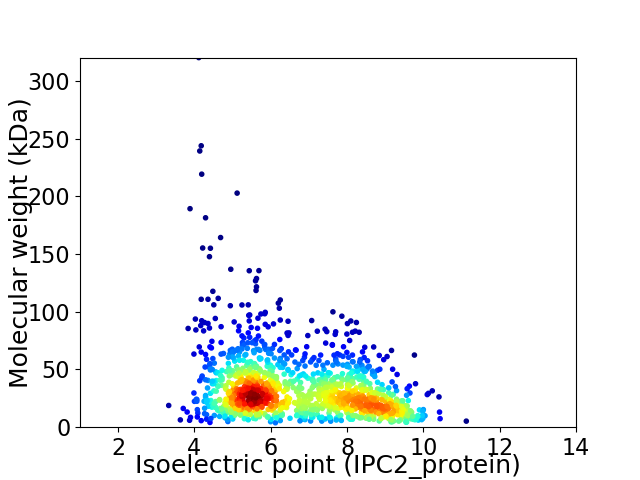
Virtual 2D-PAGE plot for 1300 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R5W5F6|R5W5F6_9DELT Lon protease OS=Corallococcus sp. CAG:1435 OX=1262867 GN=lon PE=2 SV=1
MM1 pKa = 7.63PAGNVTATANYY12 pKa = 9.74EE13 pKa = 4.25FIDD16 pKa = 3.6YY17 pKa = 8.44TVTVNGGKK25 pKa = 9.73AQVNDD30 pKa = 4.22GVASASVKK38 pKa = 10.13AHH40 pKa = 6.18YY41 pKa = 10.52GDD43 pKa = 3.95TVTITAEE50 pKa = 4.07VPVGKK55 pKa = 10.07EE56 pKa = 3.39FVMWSFNGLVPDD68 pKa = 5.49GLTLTNKK75 pKa = 9.79EE76 pKa = 4.28LTFTMPANAVTATAEE91 pKa = 3.96LRR93 pKa = 11.84DD94 pKa = 3.9LVKK97 pKa = 10.93SSVTVQGGVAQVGDD111 pKa = 4.28GEE113 pKa = 5.16LEE115 pKa = 4.21TSLEE119 pKa = 4.34AYY121 pKa = 10.5CGDD124 pKa = 3.75TVTLTAYY131 pKa = 10.33QYY133 pKa = 10.25TDD135 pKa = 2.8KK136 pKa = 11.17TFVKK140 pKa = 8.43WTIEE144 pKa = 4.5GIDD147 pKa = 3.75TEE149 pKa = 5.24GLDD152 pKa = 3.36LTQRR156 pKa = 11.84EE157 pKa = 4.69LTFTMPEE164 pKa = 3.79NDD166 pKa = 3.13VTATAEE172 pKa = 4.15FEE174 pKa = 4.31NVPTEE179 pKa = 3.96LTFEE183 pKa = 4.72GNTATSGEE191 pKa = 4.25LWYY194 pKa = 11.18VNYY197 pKa = 10.09DD198 pKa = 3.43SKK200 pKa = 11.59SFRR203 pKa = 11.84LPLGSGQDD211 pKa = 3.18SCTYY215 pKa = 10.05TIEE218 pKa = 4.31LSNSAFTFKK227 pKa = 10.99VYY229 pKa = 10.69GSDD232 pKa = 3.5GEE234 pKa = 4.49VYY236 pKa = 9.98IDD238 pKa = 3.73EE239 pKa = 4.58NGEE242 pKa = 4.23GVKK245 pKa = 10.58SITIDD250 pKa = 3.41RR251 pKa = 11.84EE252 pKa = 3.99DD253 pKa = 3.57NYY255 pKa = 10.72TIEE258 pKa = 4.45VINTGTAANCTLTVTQII275 pKa = 3.31
MM1 pKa = 7.63PAGNVTATANYY12 pKa = 9.74EE13 pKa = 4.25FIDD16 pKa = 3.6YY17 pKa = 8.44TVTVNGGKK25 pKa = 9.73AQVNDD30 pKa = 4.22GVASASVKK38 pKa = 10.13AHH40 pKa = 6.18YY41 pKa = 10.52GDD43 pKa = 3.95TVTITAEE50 pKa = 4.07VPVGKK55 pKa = 10.07EE56 pKa = 3.39FVMWSFNGLVPDD68 pKa = 5.49GLTLTNKK75 pKa = 9.79EE76 pKa = 4.28LTFTMPANAVTATAEE91 pKa = 3.96LRR93 pKa = 11.84DD94 pKa = 3.9LVKK97 pKa = 10.93SSVTVQGGVAQVGDD111 pKa = 4.28GEE113 pKa = 5.16LEE115 pKa = 4.21TSLEE119 pKa = 4.34AYY121 pKa = 10.5CGDD124 pKa = 3.75TVTLTAYY131 pKa = 10.33QYY133 pKa = 10.25TDD135 pKa = 2.8KK136 pKa = 11.17TFVKK140 pKa = 8.43WTIEE144 pKa = 4.5GIDD147 pKa = 3.75TEE149 pKa = 5.24GLDD152 pKa = 3.36LTQRR156 pKa = 11.84EE157 pKa = 4.69LTFTMPEE164 pKa = 3.79NDD166 pKa = 3.13VTATAEE172 pKa = 4.15FEE174 pKa = 4.31NVPTEE179 pKa = 3.96LTFEE183 pKa = 4.72GNTATSGEE191 pKa = 4.25LWYY194 pKa = 11.18VNYY197 pKa = 10.09DD198 pKa = 3.43SKK200 pKa = 11.59SFRR203 pKa = 11.84LPLGSGQDD211 pKa = 3.18SCTYY215 pKa = 10.05TIEE218 pKa = 4.31LSNSAFTFKK227 pKa = 10.99VYY229 pKa = 10.69GSDD232 pKa = 3.5GEE234 pKa = 4.49VYY236 pKa = 9.98IDD238 pKa = 3.73EE239 pKa = 4.58NGEE242 pKa = 4.23GVKK245 pKa = 10.58SITIDD250 pKa = 3.41RR251 pKa = 11.84EE252 pKa = 3.99DD253 pKa = 3.57NYY255 pKa = 10.72TIEE258 pKa = 4.45VINTGTAANCTLTVTQII275 pKa = 3.31
Molecular weight: 29.63 kDa
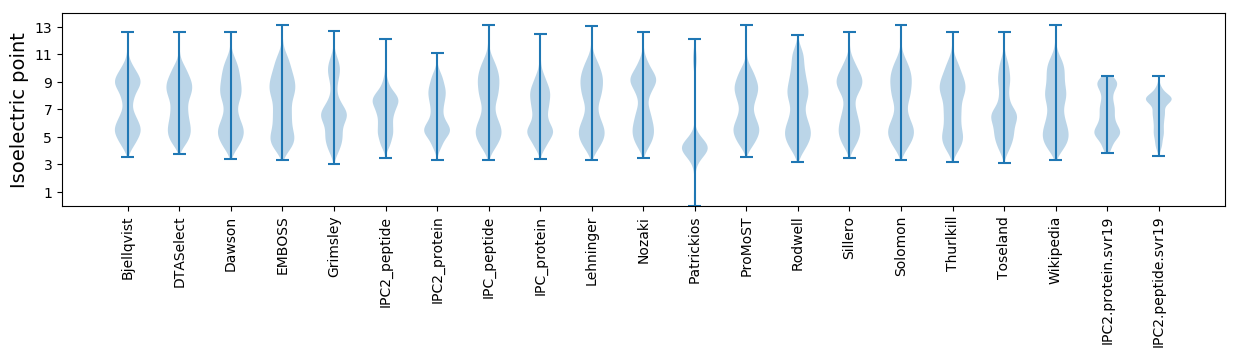
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R5WHI6|R5WHI6_9DELT Cation-transporting ATPase P-type OS=Corallococcus sp. CAG:1435 OX=1262867 GN=BN495_00525 PE=4 SV=1
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.67QPKK8 pKa = 9.11KK9 pKa = 7.52RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.17VHH16 pKa = 5.94GFRR19 pKa = 11.84ARR21 pKa = 11.84MKK23 pKa = 7.97TTGGRR28 pKa = 11.84RR29 pKa = 11.84TLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84NKK37 pKa = 9.66GRR39 pKa = 11.84HH40 pKa = 5.02SLSAA44 pKa = 3.67
MM1 pKa = 7.36KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 9.67QPKK8 pKa = 9.11KK9 pKa = 7.52RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.17VHH16 pKa = 5.94GFRR19 pKa = 11.84ARR21 pKa = 11.84MKK23 pKa = 7.97TTGGRR28 pKa = 11.84RR29 pKa = 11.84TLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84NKK37 pKa = 9.66GRR39 pKa = 11.84HH40 pKa = 5.02SLSAA44 pKa = 3.67
Molecular weight: 5.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
407049 |
34 |
2964 |
313.1 |
34.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.63 ± 0.07 | 2.005 ± 0.036 |
5.587 ± 0.052 | 5.967 ± 0.066 |
4.763 ± 0.058 | 6.681 ± 0.067 |
1.636 ± 0.037 | 5.935 ± 0.068 |
6.908 ± 0.074 | 9.121 ± 0.084 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.376 ± 0.037 | 4.909 ± 0.047 |
3.028 ± 0.033 | 3.854 ± 0.053 |
4.074 ± 0.068 | 6.062 ± 0.058 |
6.062 ± 0.1 | 8.654 ± 0.067 |
0.789 ± 0.024 | 3.952 ± 0.055 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
