
Torque teno virus (isolate Human/Finland/Hel32/2002) (TTV) (Torque teno virus genotype 6)
Taxonomy: Viruses; Anelloviridae; Alphatorquevirus
Average proteome isoelectric point is 8.44
Get precalculated fractions of proteins
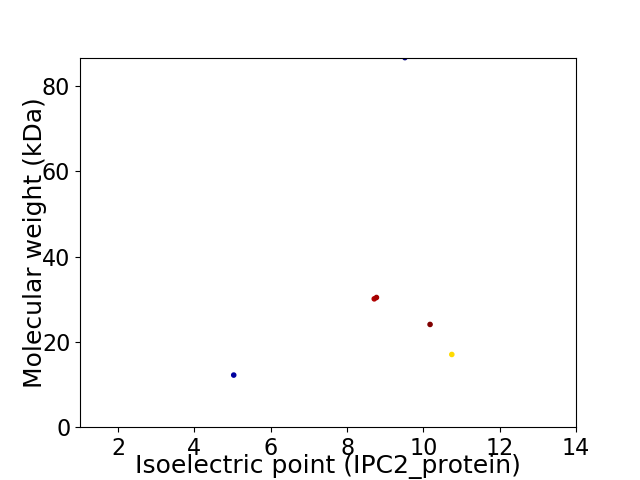
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>sp|A7XCE1|ORF12_TTVV3 ORF1/2 protein OS=Torque teno virus (isolate Human/Finland/Hel32/2002) OX=687342 GN=ORF1/2 PE=4 SV=1
MM1 pKa = 7.17WQPPTQNGTQLEE13 pKa = 4.44RR14 pKa = 11.84HH15 pKa = 5.12WFEE18 pKa = 4.58SVWRR22 pKa = 11.84SHH24 pKa = 7.0AAFCSCGDD32 pKa = 4.27CIGHH36 pKa = 6.57LQHH39 pKa = 7.35LATNLGRR46 pKa = 11.84PPAPQPPRR54 pKa = 11.84DD55 pKa = 3.49QHH57 pKa = 6.07PPHH60 pKa = 6.85IRR62 pKa = 11.84GLPALPAPPSNRR74 pKa = 11.84NSWPGTGGDD83 pKa = 3.68AAGGEE88 pKa = 4.51AGGSRR93 pKa = 11.84GAGDD97 pKa = 3.8GGDD100 pKa = 4.01GEE102 pKa = 5.38LADD105 pKa = 5.23EE106 pKa = 5.14DD107 pKa = 4.53LLDD110 pKa = 5.37AIALAAEE117 pKa = 4.3
MM1 pKa = 7.17WQPPTQNGTQLEE13 pKa = 4.44RR14 pKa = 11.84HH15 pKa = 5.12WFEE18 pKa = 4.58SVWRR22 pKa = 11.84SHH24 pKa = 7.0AAFCSCGDD32 pKa = 4.27CIGHH36 pKa = 6.57LQHH39 pKa = 7.35LATNLGRR46 pKa = 11.84PPAPQPPRR54 pKa = 11.84DD55 pKa = 3.49QHH57 pKa = 6.07PPHH60 pKa = 6.85IRR62 pKa = 11.84GLPALPAPPSNRR74 pKa = 11.84NSWPGTGGDD83 pKa = 3.68AAGGEE88 pKa = 4.51AGGSRR93 pKa = 11.84GAGDD97 pKa = 3.8GGDD100 pKa = 4.01GEE102 pKa = 5.38LADD105 pKa = 5.23EE106 pKa = 5.14DD107 pKa = 4.53LLDD110 pKa = 5.37AIALAAEE117 pKa = 4.3
Molecular weight: 12.23 kDa
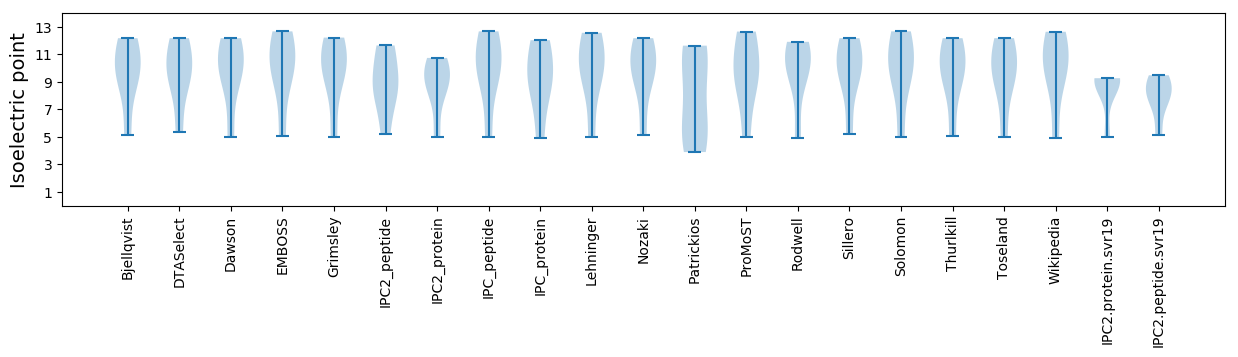
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|A7XCE8|ORF11_TTVV3 ORF1/1 protein OS=Torque teno virus (isolate Human/Finland/Hel32/2002) OX=687342 GN=ORF1/1 PE=4 SV=1
MM1 pKa = 7.26AWYY4 pKa = 7.29WWRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84GWWKK16 pKa = 8.57PRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84WRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84ARR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84GPARR34 pKa = 11.84RR35 pKa = 11.84HH36 pKa = 4.81RR37 pKa = 11.84ARR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84VRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84GRR48 pKa = 11.84WRR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84YY53 pKa = 9.42RR54 pKa = 11.84RR55 pKa = 11.84WRR57 pKa = 11.84RR58 pKa = 11.84RR59 pKa = 11.84GGRR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84HH65 pKa = 5.47RR66 pKa = 11.84KK67 pKa = 9.32KK68 pKa = 10.97LIIKK72 pKa = 8.44QWQPNFIRR80 pKa = 11.84HH81 pKa = 6.18CYY83 pKa = 9.05IIGYY87 pKa = 8.03MPLIICGEE95 pKa = 4.1NTFSHH100 pKa = 6.82NYY102 pKa = 7.27ATHH105 pKa = 6.53SDD107 pKa = 3.86DD108 pKa = 3.92MLSTGPYY115 pKa = 8.57GGGMTTTKK123 pKa = 9.14FTLRR127 pKa = 11.84ILFDD131 pKa = 3.75EE132 pKa = 4.62YY133 pKa = 10.98QRR135 pKa = 11.84HH136 pKa = 5.46LNFWTVSNQDD146 pKa = 3.14LDD148 pKa = 3.75LARR151 pKa = 11.84YY152 pKa = 9.04LGTKK156 pKa = 9.84IIFFRR161 pKa = 11.84HH162 pKa = 4.71PTVDD166 pKa = 3.82FVVQIHH172 pKa = 5.65TQPPFQDD179 pKa = 3.69TEE181 pKa = 3.84ITAPSIHH188 pKa = 7.21PGMLILSKK196 pKa = 10.66KK197 pKa = 9.88HH198 pKa = 6.28ILIPSLKK205 pKa = 8.33TRR207 pKa = 11.84PSKK210 pKa = 10.15KK211 pKa = 8.85HH212 pKa = 4.02YY213 pKa = 9.27VKK215 pKa = 10.69VRR217 pKa = 11.84VGAPRR222 pKa = 11.84LFQDD226 pKa = 2.36KK227 pKa = 9.6WYY229 pKa = 8.57PQSEE233 pKa = 4.59LCDD236 pKa = 3.43VTLLVIYY243 pKa = 8.35ATACDD248 pKa = 3.69LQYY251 pKa = 11.21PFGSPQTDD259 pKa = 3.27NVCVNFQILGQPYY272 pKa = 8.15YY273 pKa = 10.86QHH275 pKa = 7.12LKK277 pKa = 8.08TALGLTEE284 pKa = 3.69KK285 pKa = 8.12TTYY288 pKa = 9.8EE289 pKa = 3.63NHH291 pKa = 6.22YY292 pKa = 10.68KK293 pKa = 10.63NNLYY297 pKa = 10.7KK298 pKa = 10.59KK299 pKa = 9.34IKK301 pKa = 10.08FYY303 pKa = 10.71NTTEE307 pKa = 4.23TIAQLKK313 pKa = 8.97PLVDD317 pKa = 3.27ATTNQTWSHH326 pKa = 4.94YY327 pKa = 10.28VNPNKK332 pKa = 9.83LTTTPTSEE340 pKa = 3.68ITHH343 pKa = 6.32NNTWYY348 pKa = 10.65RR349 pKa = 11.84GNAYY353 pKa = 10.03NDD355 pKa = 4.37KK356 pKa = 9.67ITDD359 pKa = 3.49LPEE362 pKa = 4.02IVKK365 pKa = 10.39KK366 pKa = 10.57SYY368 pKa = 11.28YY369 pKa = 9.99KK370 pKa = 10.05ATEE373 pKa = 3.73LAIPEE378 pKa = 4.25AVKK381 pKa = 9.0PTTDD385 pKa = 3.1LFEE388 pKa = 4.19YY389 pKa = 10.4HH390 pKa = 6.57AGIYY394 pKa = 10.25SSIFLSPGRR403 pKa = 11.84AYY405 pKa = 10.82FEE407 pKa = 4.53TPGAYY412 pKa = 9.41QDD414 pKa = 4.04IIYY417 pKa = 10.97NPFTDD422 pKa = 3.83KK423 pKa = 11.45GIGNIVWIDD432 pKa = 3.39WLSKK436 pKa = 10.4SDD438 pKa = 3.3AVYY441 pKa = 10.69SEE443 pKa = 4.37KK444 pKa = 10.29QSKK447 pKa = 10.49CGIFDD452 pKa = 4.07LPLWAAFFGYY462 pKa = 10.98AEE464 pKa = 4.53FCSKK468 pKa = 9.84STGDD472 pKa = 3.11TAIAYY477 pKa = 8.15NSRR480 pKa = 11.84VCVRR484 pKa = 11.84CPYY487 pKa = 10.06TEE489 pKa = 4.17PQLLNHH495 pKa = 6.51NNPSQGYY502 pKa = 8.23VFYY505 pKa = 10.59SYY507 pKa = 11.73NFGKK511 pKa = 10.8GKK513 pKa = 9.25MPGGSSQVPIRR524 pKa = 11.84MRR526 pKa = 11.84WKK528 pKa = 8.49WYY530 pKa = 9.61VCMFHH535 pKa = 6.86QLEE538 pKa = 4.23VMEE541 pKa = 5.85AICQSGPFAYY551 pKa = 10.3HH552 pKa = 6.23SDD554 pKa = 3.43EE555 pKa = 4.24KK556 pKa = 11.1KK557 pKa = 10.35AVLGIKK563 pKa = 10.31YY564 pKa = 10.32KK565 pKa = 10.67FDD567 pKa = 3.7WKK569 pKa = 10.45WGGNPISQQIVRR581 pKa = 11.84HH582 pKa = 5.54PCNGQTSSGNRR593 pKa = 11.84VPRR596 pKa = 11.84SVQAVDD602 pKa = 3.51PKK604 pKa = 10.9YY605 pKa = 11.25VSLQLVWHH613 pKa = 6.2SWDD616 pKa = 3.06FRR618 pKa = 11.84RR619 pKa = 11.84GLFGQAGIKK628 pKa = 10.19RR629 pKa = 11.84MQQEE633 pKa = 4.02SDD635 pKa = 3.62ALTLSPVHH643 pKa = 6.3RR644 pKa = 11.84PKK646 pKa = 10.63RR647 pKa = 11.84PKK649 pKa = 10.02RR650 pKa = 11.84DD651 pKa = 3.47TQVKK655 pKa = 9.68EE656 pKa = 3.76KK657 pKa = 10.22TPEE660 pKa = 3.97KK661 pKa = 10.74DD662 pKa = 2.62SDD664 pKa = 3.86SAVQLRR670 pKa = 11.84RR671 pKa = 11.84LQPWIHH677 pKa = 6.47SSQEE681 pKa = 4.06TKK683 pKa = 10.81DD684 pKa = 3.62EE685 pKa = 4.14EE686 pKa = 4.39EE687 pKa = 5.02EE688 pKa = 4.18IPEE691 pKa = 4.55GPVQEE696 pKa = 4.76QLLQQLQQQRR706 pKa = 11.84LLRR709 pKa = 11.84VQLEE713 pKa = 4.54SIAQEE718 pKa = 3.8VLKK721 pKa = 10.51IRR723 pKa = 11.84RR724 pKa = 11.84GHH726 pKa = 5.64SLHH729 pKa = 6.95PLLSSHH735 pKa = 6.96AA736 pKa = 4.08
MM1 pKa = 7.26AWYY4 pKa = 7.29WWRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84GWWKK16 pKa = 8.57PRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84RR21 pKa = 11.84WRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84ARR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84GPARR34 pKa = 11.84RR35 pKa = 11.84HH36 pKa = 4.81RR37 pKa = 11.84ARR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84VRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84GRR48 pKa = 11.84WRR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84YY53 pKa = 9.42RR54 pKa = 11.84RR55 pKa = 11.84WRR57 pKa = 11.84RR58 pKa = 11.84RR59 pKa = 11.84GGRR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84HH65 pKa = 5.47RR66 pKa = 11.84KK67 pKa = 9.32KK68 pKa = 10.97LIIKK72 pKa = 8.44QWQPNFIRR80 pKa = 11.84HH81 pKa = 6.18CYY83 pKa = 9.05IIGYY87 pKa = 8.03MPLIICGEE95 pKa = 4.1NTFSHH100 pKa = 6.82NYY102 pKa = 7.27ATHH105 pKa = 6.53SDD107 pKa = 3.86DD108 pKa = 3.92MLSTGPYY115 pKa = 8.57GGGMTTTKK123 pKa = 9.14FTLRR127 pKa = 11.84ILFDD131 pKa = 3.75EE132 pKa = 4.62YY133 pKa = 10.98QRR135 pKa = 11.84HH136 pKa = 5.46LNFWTVSNQDD146 pKa = 3.14LDD148 pKa = 3.75LARR151 pKa = 11.84YY152 pKa = 9.04LGTKK156 pKa = 9.84IIFFRR161 pKa = 11.84HH162 pKa = 4.71PTVDD166 pKa = 3.82FVVQIHH172 pKa = 5.65TQPPFQDD179 pKa = 3.69TEE181 pKa = 3.84ITAPSIHH188 pKa = 7.21PGMLILSKK196 pKa = 10.66KK197 pKa = 9.88HH198 pKa = 6.28ILIPSLKK205 pKa = 8.33TRR207 pKa = 11.84PSKK210 pKa = 10.15KK211 pKa = 8.85HH212 pKa = 4.02YY213 pKa = 9.27VKK215 pKa = 10.69VRR217 pKa = 11.84VGAPRR222 pKa = 11.84LFQDD226 pKa = 2.36KK227 pKa = 9.6WYY229 pKa = 8.57PQSEE233 pKa = 4.59LCDD236 pKa = 3.43VTLLVIYY243 pKa = 8.35ATACDD248 pKa = 3.69LQYY251 pKa = 11.21PFGSPQTDD259 pKa = 3.27NVCVNFQILGQPYY272 pKa = 8.15YY273 pKa = 10.86QHH275 pKa = 7.12LKK277 pKa = 8.08TALGLTEE284 pKa = 3.69KK285 pKa = 8.12TTYY288 pKa = 9.8EE289 pKa = 3.63NHH291 pKa = 6.22YY292 pKa = 10.68KK293 pKa = 10.63NNLYY297 pKa = 10.7KK298 pKa = 10.59KK299 pKa = 9.34IKK301 pKa = 10.08FYY303 pKa = 10.71NTTEE307 pKa = 4.23TIAQLKK313 pKa = 8.97PLVDD317 pKa = 3.27ATTNQTWSHH326 pKa = 4.94YY327 pKa = 10.28VNPNKK332 pKa = 9.83LTTTPTSEE340 pKa = 3.68ITHH343 pKa = 6.32NNTWYY348 pKa = 10.65RR349 pKa = 11.84GNAYY353 pKa = 10.03NDD355 pKa = 4.37KK356 pKa = 9.67ITDD359 pKa = 3.49LPEE362 pKa = 4.02IVKK365 pKa = 10.39KK366 pKa = 10.57SYY368 pKa = 11.28YY369 pKa = 9.99KK370 pKa = 10.05ATEE373 pKa = 3.73LAIPEE378 pKa = 4.25AVKK381 pKa = 9.0PTTDD385 pKa = 3.1LFEE388 pKa = 4.19YY389 pKa = 10.4HH390 pKa = 6.57AGIYY394 pKa = 10.25SSIFLSPGRR403 pKa = 11.84AYY405 pKa = 10.82FEE407 pKa = 4.53TPGAYY412 pKa = 9.41QDD414 pKa = 4.04IIYY417 pKa = 10.97NPFTDD422 pKa = 3.83KK423 pKa = 11.45GIGNIVWIDD432 pKa = 3.39WLSKK436 pKa = 10.4SDD438 pKa = 3.3AVYY441 pKa = 10.69SEE443 pKa = 4.37KK444 pKa = 10.29QSKK447 pKa = 10.49CGIFDD452 pKa = 4.07LPLWAAFFGYY462 pKa = 10.98AEE464 pKa = 4.53FCSKK468 pKa = 9.84STGDD472 pKa = 3.11TAIAYY477 pKa = 8.15NSRR480 pKa = 11.84VCVRR484 pKa = 11.84CPYY487 pKa = 10.06TEE489 pKa = 4.17PQLLNHH495 pKa = 6.51NNPSQGYY502 pKa = 8.23VFYY505 pKa = 10.59SYY507 pKa = 11.73NFGKK511 pKa = 10.8GKK513 pKa = 9.25MPGGSSQVPIRR524 pKa = 11.84MRR526 pKa = 11.84WKK528 pKa = 8.49WYY530 pKa = 9.61VCMFHH535 pKa = 6.86QLEE538 pKa = 4.23VMEE541 pKa = 5.85AICQSGPFAYY551 pKa = 10.3HH552 pKa = 6.23SDD554 pKa = 3.43EE555 pKa = 4.24KK556 pKa = 11.1KK557 pKa = 10.35AVLGIKK563 pKa = 10.31YY564 pKa = 10.32KK565 pKa = 10.67FDD567 pKa = 3.7WKK569 pKa = 10.45WGGNPISQQIVRR581 pKa = 11.84HH582 pKa = 5.54PCNGQTSSGNRR593 pKa = 11.84VPRR596 pKa = 11.84SVQAVDD602 pKa = 3.51PKK604 pKa = 10.9YY605 pKa = 11.25VSLQLVWHH613 pKa = 6.2SWDD616 pKa = 3.06FRR618 pKa = 11.84RR619 pKa = 11.84GLFGQAGIKK628 pKa = 10.19RR629 pKa = 11.84MQQEE633 pKa = 4.02SDD635 pKa = 3.62ALTLSPVHH643 pKa = 6.3RR644 pKa = 11.84PKK646 pKa = 10.63RR647 pKa = 11.84PKK649 pKa = 10.02RR650 pKa = 11.84DD651 pKa = 3.47TQVKK655 pKa = 9.68EE656 pKa = 3.76KK657 pKa = 10.22TPEE660 pKa = 3.97KK661 pKa = 10.74DD662 pKa = 2.62SDD664 pKa = 3.86SAVQLRR670 pKa = 11.84RR671 pKa = 11.84LQPWIHH677 pKa = 6.47SSQEE681 pKa = 4.06TKK683 pKa = 10.81DD684 pKa = 3.62EE685 pKa = 4.14EE686 pKa = 4.39EE687 pKa = 5.02EE688 pKa = 4.18IPEE691 pKa = 4.55GPVQEE696 pKa = 4.76QLLQQLQQQRR706 pKa = 11.84LLRR709 pKa = 11.84VQLEE713 pKa = 4.54SIAQEE718 pKa = 3.8VLKK721 pKa = 10.51IRR723 pKa = 11.84RR724 pKa = 11.84GHH726 pKa = 5.64SLHH729 pKa = 6.95PLLSSHH735 pKa = 6.96AA736 pKa = 4.08
Molecular weight: 86.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1750 |
117 |
736 |
291.7 |
33.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.686 ± 1.203 | 1.6 ± 0.299 |
4.286 ± 0.407 | 4.286 ± 0.359 |
2.686 ± 0.654 | 7.2 ± 1.129 |
3.429 ± 0.307 | 3.714 ± 1.097 |
5.6 ± 0.986 | 6.743 ± 0.779 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.2 ± 0.083 | 3.429 ± 0.383 |
8.571 ± 1.562 | 6.229 ± 0.517 |
10.914 ± 2.183 | 8.057 ± 1.877 |
5.657 ± 0.561 | 3.429 ± 0.975 |
3.029 ± 0.401 | 3.257 ± 1.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
