
Hubei dimarhabdovirus virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
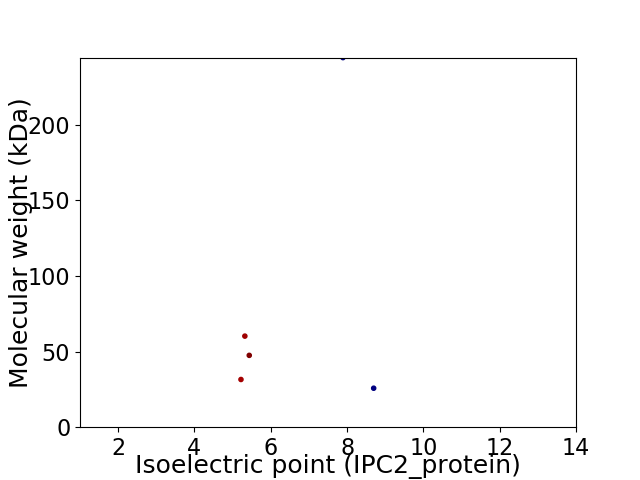
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KMW1|A0A1L3KMW1_9VIRU Putative glycoprotein OS=Hubei dimarhabdovirus virus 2 OX=1922867 PE=4 SV=1
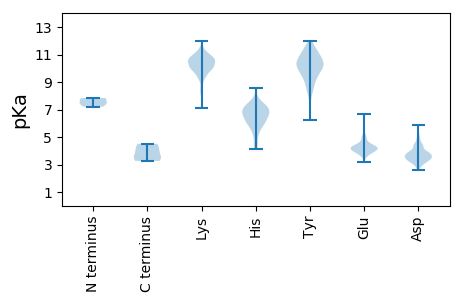
MM1 pKa = 7.41YY2 pKa = 10.33SLQGGLTDD10 pKa = 3.43VKK12 pKa = 10.72EE13 pKa = 4.37VLNSNPNYY21 pKa = 9.94LRR23 pKa = 11.84KK24 pKa = 9.67SYY26 pKa = 11.01QMSTDD31 pKa = 3.3EE32 pKa = 4.95EE33 pKa = 5.08KK34 pKa = 11.29YY35 pKa = 9.47STPPLSASLISEE47 pKa = 4.16DD48 pKa = 4.78CKK50 pKa = 11.37NLTDD54 pKa = 3.61QWSEE58 pKa = 3.88APIIPTDD65 pKa = 3.41GMVDD69 pKa = 3.04QGLILEE75 pKa = 4.33MEE77 pKa = 4.26IDD79 pKa = 3.72NEE81 pKa = 3.94KK82 pKa = 10.96DD83 pKa = 2.93HH84 pKa = 7.09ASRR87 pKa = 11.84IEE89 pKa = 3.86MSKK92 pKa = 10.97LKK94 pKa = 9.84TPPQDD99 pKa = 3.34DD100 pKa = 3.66QRR102 pKa = 11.84NEE104 pKa = 3.51TSKK107 pKa = 10.52RR108 pKa = 11.84SRR110 pKa = 11.84IITPEE115 pKa = 3.64EE116 pKa = 4.15DD117 pKa = 2.86KK118 pKa = 11.18WNIAIDD124 pKa = 3.76MGSLSLQDD132 pKa = 3.27KK133 pKa = 10.07MSFKK137 pKa = 10.69EE138 pKa = 3.97AFKK141 pKa = 10.65KK142 pKa = 10.3HH143 pKa = 4.2VDD145 pKa = 3.46AFVKK149 pKa = 10.55GCRR152 pKa = 11.84IPLNINYY159 pKa = 9.53DD160 pKa = 3.92IEE162 pKa = 4.78PMDD165 pKa = 3.52IQEE168 pKa = 4.15SKK170 pKa = 10.6IRR172 pKa = 11.84VIKK175 pKa = 9.86TSSPSEE181 pKa = 4.13GDD183 pKa = 3.61SMAMAQKK190 pKa = 10.63PPDD193 pKa = 4.01PEE195 pKa = 5.92LNDD198 pKa = 3.91TQKK201 pKa = 11.51KK202 pKa = 10.12LYY204 pKa = 10.51SLLLQGMEE212 pKa = 5.02LPLLSNPKK220 pKa = 9.87RR221 pKa = 11.84YY222 pKa = 8.8FVLKK226 pKa = 10.03IGKK229 pKa = 9.48DD230 pKa = 3.41EE231 pKa = 4.51FEE233 pKa = 4.68GNAVLDD239 pKa = 4.28VIIRR243 pKa = 11.84SPDD246 pKa = 3.14LDD248 pKa = 3.21IRR250 pKa = 11.84SLLRR254 pKa = 11.84RR255 pKa = 11.84VISISQQKK263 pKa = 10.61KK264 pKa = 9.57MILNLYY270 pKa = 9.22KK271 pKa = 9.82WDD273 pKa = 3.68AKK275 pKa = 10.55II276 pKa = 4.26
MM1 pKa = 7.41YY2 pKa = 10.33SLQGGLTDD10 pKa = 3.43VKK12 pKa = 10.72EE13 pKa = 4.37VLNSNPNYY21 pKa = 9.94LRR23 pKa = 11.84KK24 pKa = 9.67SYY26 pKa = 11.01QMSTDD31 pKa = 3.3EE32 pKa = 4.95EE33 pKa = 5.08KK34 pKa = 11.29YY35 pKa = 9.47STPPLSASLISEE47 pKa = 4.16DD48 pKa = 4.78CKK50 pKa = 11.37NLTDD54 pKa = 3.61QWSEE58 pKa = 3.88APIIPTDD65 pKa = 3.41GMVDD69 pKa = 3.04QGLILEE75 pKa = 4.33MEE77 pKa = 4.26IDD79 pKa = 3.72NEE81 pKa = 3.94KK82 pKa = 10.96DD83 pKa = 2.93HH84 pKa = 7.09ASRR87 pKa = 11.84IEE89 pKa = 3.86MSKK92 pKa = 10.97LKK94 pKa = 9.84TPPQDD99 pKa = 3.34DD100 pKa = 3.66QRR102 pKa = 11.84NEE104 pKa = 3.51TSKK107 pKa = 10.52RR108 pKa = 11.84SRR110 pKa = 11.84IITPEE115 pKa = 3.64EE116 pKa = 4.15DD117 pKa = 2.86KK118 pKa = 11.18WNIAIDD124 pKa = 3.76MGSLSLQDD132 pKa = 3.27KK133 pKa = 10.07MSFKK137 pKa = 10.69EE138 pKa = 3.97AFKK141 pKa = 10.65KK142 pKa = 10.3HH143 pKa = 4.2VDD145 pKa = 3.46AFVKK149 pKa = 10.55GCRR152 pKa = 11.84IPLNINYY159 pKa = 9.53DD160 pKa = 3.92IEE162 pKa = 4.78PMDD165 pKa = 3.52IQEE168 pKa = 4.15SKK170 pKa = 10.6IRR172 pKa = 11.84VIKK175 pKa = 9.86TSSPSEE181 pKa = 4.13GDD183 pKa = 3.61SMAMAQKK190 pKa = 10.63PPDD193 pKa = 4.01PEE195 pKa = 5.92LNDD198 pKa = 3.91TQKK201 pKa = 11.51KK202 pKa = 10.12LYY204 pKa = 10.51SLLLQGMEE212 pKa = 5.02LPLLSNPKK220 pKa = 9.87RR221 pKa = 11.84YY222 pKa = 8.8FVLKK226 pKa = 10.03IGKK229 pKa = 9.48DD230 pKa = 3.41EE231 pKa = 4.51FEE233 pKa = 4.68GNAVLDD239 pKa = 4.28VIIRR243 pKa = 11.84SPDD246 pKa = 3.14LDD248 pKa = 3.21IRR250 pKa = 11.84SLLRR254 pKa = 11.84RR255 pKa = 11.84VISISQQKK263 pKa = 10.61KK264 pKa = 9.57MILNLYY270 pKa = 9.22KK271 pKa = 9.82WDD273 pKa = 3.68AKK275 pKa = 10.55II276 pKa = 4.26
Molecular weight: 31.54 kDa
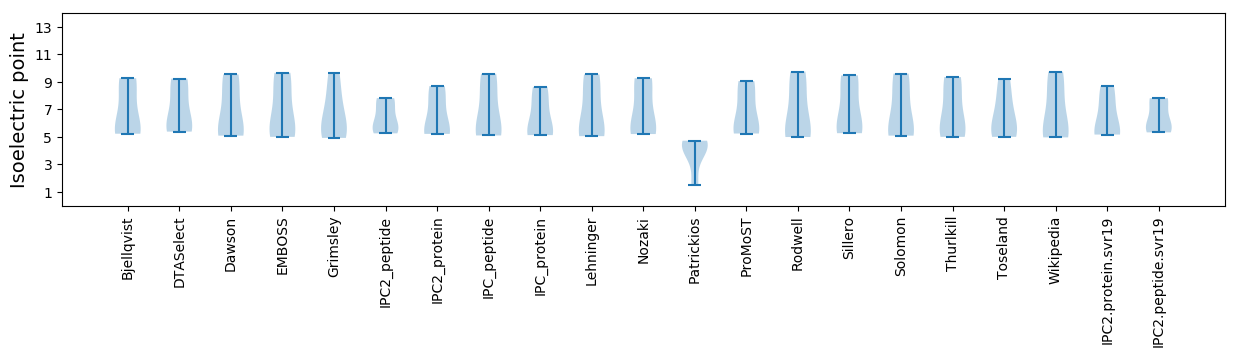
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KMT3|A0A1L3KMT3_9VIRU Uncharacterized protein OS=Hubei dimarhabdovirus virus 2 OX=1922867 PE=4 SV=1
MM1 pKa = 7.2NRR3 pKa = 11.84LFKK6 pKa = 10.78SPRR9 pKa = 11.84KK10 pKa = 9.97RR11 pKa = 11.84KK12 pKa = 8.34DD13 pKa = 3.23TKK15 pKa = 10.8SSSSSDD21 pKa = 3.3SSSGSLSEE29 pKa = 4.92LPISVWIPPTSTGPVMEE46 pKa = 5.71DD47 pKa = 2.97YY48 pKa = 10.35WIRR51 pKa = 11.84PTSPPVSIVFDD62 pKa = 3.48SHH64 pKa = 5.49IVRR67 pKa = 11.84YY68 pKa = 10.01VKK70 pKa = 10.76LNLAITSNKK79 pKa = 8.89PIKK82 pKa = 10.63NHH84 pKa = 6.98DD85 pKa = 3.52EE86 pKa = 4.42MINMLSLIIDD96 pKa = 4.5HH97 pKa = 5.91YY98 pKa = 10.85QGPIVLKK105 pKa = 9.87PVILALYY112 pKa = 8.42GAAGMSLKK120 pKa = 10.13LDD122 pKa = 3.09RR123 pKa = 11.84GMRR126 pKa = 11.84SDD128 pKa = 3.64YY129 pKa = 10.93KK130 pKa = 10.76YY131 pKa = 10.99LSRR134 pKa = 11.84LDD136 pKa = 3.87QILMFHH142 pKa = 6.38TKK144 pKa = 9.95NRR146 pKa = 11.84DD147 pKa = 3.32YY148 pKa = 11.45SEE150 pKa = 4.99EE151 pKa = 3.73ICDD154 pKa = 3.73INWIKK159 pKa = 10.08TSSLGGNRR167 pKa = 11.84YY168 pKa = 9.89SIQYY172 pKa = 9.37SLTMEE177 pKa = 4.05KK178 pKa = 10.58SNRR181 pKa = 11.84KK182 pKa = 9.53GNDD185 pKa = 2.86FFSHH189 pKa = 4.49YY190 pKa = 9.84RR191 pKa = 11.84KK192 pKa = 10.09YY193 pKa = 11.11SRR195 pKa = 11.84TEE197 pKa = 3.2KK198 pKa = 10.5SMSMEE203 pKa = 4.13VIKK206 pKa = 10.66EE207 pKa = 3.65ILNIDD212 pKa = 3.64RR213 pKa = 11.84MEE215 pKa = 4.65LEE217 pKa = 3.89SDD219 pKa = 3.46TAVFLL224 pKa = 4.51
MM1 pKa = 7.2NRR3 pKa = 11.84LFKK6 pKa = 10.78SPRR9 pKa = 11.84KK10 pKa = 9.97RR11 pKa = 11.84KK12 pKa = 8.34DD13 pKa = 3.23TKK15 pKa = 10.8SSSSSDD21 pKa = 3.3SSSGSLSEE29 pKa = 4.92LPISVWIPPTSTGPVMEE46 pKa = 5.71DD47 pKa = 2.97YY48 pKa = 10.35WIRR51 pKa = 11.84PTSPPVSIVFDD62 pKa = 3.48SHH64 pKa = 5.49IVRR67 pKa = 11.84YY68 pKa = 10.01VKK70 pKa = 10.76LNLAITSNKK79 pKa = 8.89PIKK82 pKa = 10.63NHH84 pKa = 6.98DD85 pKa = 3.52EE86 pKa = 4.42MINMLSLIIDD96 pKa = 4.5HH97 pKa = 5.91YY98 pKa = 10.85QGPIVLKK105 pKa = 9.87PVILALYY112 pKa = 8.42GAAGMSLKK120 pKa = 10.13LDD122 pKa = 3.09RR123 pKa = 11.84GMRR126 pKa = 11.84SDD128 pKa = 3.64YY129 pKa = 10.93KK130 pKa = 10.76YY131 pKa = 10.99LSRR134 pKa = 11.84LDD136 pKa = 3.87QILMFHH142 pKa = 6.38TKK144 pKa = 9.95NRR146 pKa = 11.84DD147 pKa = 3.32YY148 pKa = 11.45SEE150 pKa = 4.99EE151 pKa = 3.73ICDD154 pKa = 3.73INWIKK159 pKa = 10.08TSSLGGNRR167 pKa = 11.84YY168 pKa = 9.89SIQYY172 pKa = 9.37SLTMEE177 pKa = 4.05KK178 pKa = 10.58SNRR181 pKa = 11.84KK182 pKa = 9.53GNDD185 pKa = 2.86FFSHH189 pKa = 4.49YY190 pKa = 9.84RR191 pKa = 11.84KK192 pKa = 10.09YY193 pKa = 11.11SRR195 pKa = 11.84TEE197 pKa = 3.2KK198 pKa = 10.5SMSMEE203 pKa = 4.13VIKK206 pKa = 10.66EE207 pKa = 3.65ILNIDD212 pKa = 3.64RR213 pKa = 11.84MEE215 pKa = 4.65LEE217 pKa = 3.89SDD219 pKa = 3.46TAVFLL224 pKa = 4.51
Molecular weight: 25.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3574 |
224 |
2119 |
714.8 |
81.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.302 ± 0.406 | 1.595 ± 0.376 |
6.072 ± 0.438 | 5.624 ± 0.326 |
4.001 ± 0.443 | 5.092 ± 0.28 |
2.434 ± 0.368 | 8.73 ± 0.322 |
7.387 ± 0.318 | 9.625 ± 0.52 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.882 ± 0.393 | 5.988 ± 0.31 |
4.477 ± 0.399 | 2.462 ± 0.377 |
4.393 ± 0.733 | 9.569 ± 0.645 |
5.036 ± 0.467 | 5.232 ± 0.379 |
1.623 ± 0.228 | 4.477 ± 0.295 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
