
Qinghai Lake virophage
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Preplasmiviricota; Maveriviricetes; Priklausovirales; Lavidaviridae; unclassified Lavidaviridae
Average proteome isoelectric point is 7.1
Get precalculated fractions of proteins
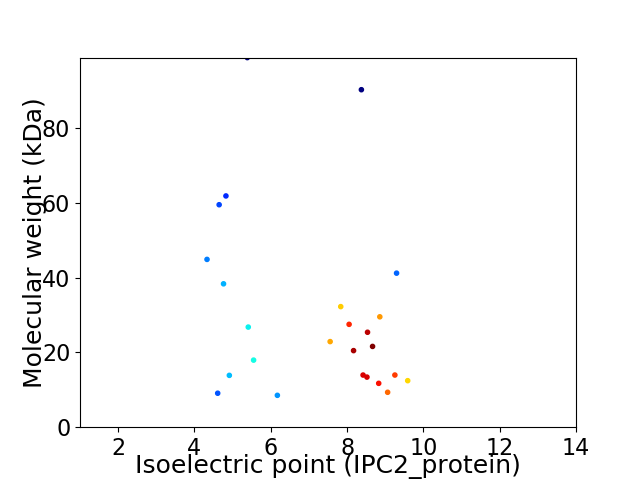
Virtual 2D-PAGE plot for 25 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0R5K6B7|A0A0R5K6B7_9VIRU Uncharacterized protein OS=Qinghai Lake virophage OX=1516115 GN=QLV_13 PE=3 SV=1
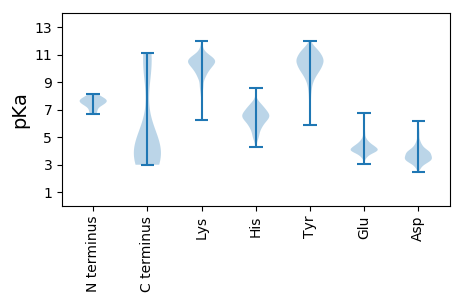
MM1 pKa = 6.98SQQVSKK7 pKa = 10.3ATTPDD12 pKa = 2.87MVYY15 pKa = 11.06YY16 pKa = 10.46DD17 pKa = 3.12IVVTNFQSITTKK29 pKa = 10.64APKK32 pKa = 10.22LRR34 pKa = 11.84FNEE37 pKa = 4.09SRR39 pKa = 11.84TNPIIDD45 pKa = 3.65KK46 pKa = 10.47TEE48 pKa = 3.99DD49 pKa = 3.61YY50 pKa = 11.1YY51 pKa = 11.32MSIVRR56 pKa = 11.84FSLDD60 pKa = 3.13TYY62 pKa = 10.54NLPNIICEE70 pKa = 4.08IQPNQADD77 pKa = 4.25PNLSIYY83 pKa = 10.86SVTLQALLPDD93 pKa = 4.03GVGGFTTTEE102 pKa = 3.8VQTFLNWIPQNRR114 pKa = 11.84DD115 pKa = 2.47IPTPLAPSLTTTGFQMTNTEE135 pKa = 4.15YY136 pKa = 10.29YY137 pKa = 9.7YY138 pKa = 10.53CYY140 pKa = 10.01QFQYY144 pKa = 10.99FLALMNQAFQTCLVDD159 pKa = 6.08LITATGGGASPINTAQQPIILWDD182 pKa = 3.76FTSNSAILNAPSNFYY197 pKa = 10.73EE198 pKa = 4.25VTIDD202 pKa = 3.44PTVNPIINIFFNKK215 pKa = 9.72PLFALFNSFLNIDD228 pKa = 4.14FGVSITNGKK237 pKa = 7.76NHH239 pKa = 6.57QIVIDD244 pKa = 4.32SYY246 pKa = 11.66LGTQTIQFPYY256 pKa = 10.66GLDD259 pKa = 3.4PEE261 pKa = 5.41LEE263 pKa = 4.32YY264 pKa = 10.51TQTFQEE270 pKa = 4.99FSTIDD275 pKa = 2.63TWTPVSSIVFTSNTIPIVSNQLSAPLIFNDD305 pKa = 4.06GQIVVGDD312 pKa = 3.78GNNCNFAQIITDD324 pKa = 4.3LEE326 pKa = 4.51TNQQVYY332 pKa = 9.71KK333 pKa = 10.13PQLLYY338 pKa = 11.19VPTAEE343 pKa = 4.22YY344 pKa = 10.68RR345 pKa = 11.84RR346 pKa = 11.84IDD348 pKa = 3.48LTGNRR353 pKa = 11.84PLTNIDD359 pKa = 3.31IEE361 pKa = 4.82VYY363 pKa = 9.64WRR365 pKa = 11.84DD366 pKa = 3.24KK367 pKa = 11.4LGGLNQLNLEE377 pKa = 4.45SGGSATIKK385 pKa = 10.56FLFEE389 pKa = 4.49KK390 pKa = 10.31KK391 pKa = 10.1DD392 pKa = 3.27KK393 pKa = 11.34LGIKK397 pKa = 10.26GYY399 pKa = 10.86
MM1 pKa = 6.98SQQVSKK7 pKa = 10.3ATTPDD12 pKa = 2.87MVYY15 pKa = 11.06YY16 pKa = 10.46DD17 pKa = 3.12IVVTNFQSITTKK29 pKa = 10.64APKK32 pKa = 10.22LRR34 pKa = 11.84FNEE37 pKa = 4.09SRR39 pKa = 11.84TNPIIDD45 pKa = 3.65KK46 pKa = 10.47TEE48 pKa = 3.99DD49 pKa = 3.61YY50 pKa = 11.1YY51 pKa = 11.32MSIVRR56 pKa = 11.84FSLDD60 pKa = 3.13TYY62 pKa = 10.54NLPNIICEE70 pKa = 4.08IQPNQADD77 pKa = 4.25PNLSIYY83 pKa = 10.86SVTLQALLPDD93 pKa = 4.03GVGGFTTTEE102 pKa = 3.8VQTFLNWIPQNRR114 pKa = 11.84DD115 pKa = 2.47IPTPLAPSLTTTGFQMTNTEE135 pKa = 4.15YY136 pKa = 10.29YY137 pKa = 9.7YY138 pKa = 10.53CYY140 pKa = 10.01QFQYY144 pKa = 10.99FLALMNQAFQTCLVDD159 pKa = 6.08LITATGGGASPINTAQQPIILWDD182 pKa = 3.76FTSNSAILNAPSNFYY197 pKa = 10.73EE198 pKa = 4.25VTIDD202 pKa = 3.44PTVNPIINIFFNKK215 pKa = 9.72PLFALFNSFLNIDD228 pKa = 4.14FGVSITNGKK237 pKa = 7.76NHH239 pKa = 6.57QIVIDD244 pKa = 4.32SYY246 pKa = 11.66LGTQTIQFPYY256 pKa = 10.66GLDD259 pKa = 3.4PEE261 pKa = 5.41LEE263 pKa = 4.32YY264 pKa = 10.51TQTFQEE270 pKa = 4.99FSTIDD275 pKa = 2.63TWTPVSSIVFTSNTIPIVSNQLSAPLIFNDD305 pKa = 4.06GQIVVGDD312 pKa = 3.78GNNCNFAQIITDD324 pKa = 4.3LEE326 pKa = 4.51TNQQVYY332 pKa = 9.71KK333 pKa = 10.13PQLLYY338 pKa = 11.19VPTAEE343 pKa = 4.22YY344 pKa = 10.68RR345 pKa = 11.84RR346 pKa = 11.84IDD348 pKa = 3.48LTGNRR353 pKa = 11.84PLTNIDD359 pKa = 3.31IEE361 pKa = 4.82VYY363 pKa = 9.64WRR365 pKa = 11.84DD366 pKa = 3.24KK367 pKa = 11.4LGGLNQLNLEE377 pKa = 4.45SGGSATIKK385 pKa = 10.56FLFEE389 pKa = 4.49KK390 pKa = 10.31KK391 pKa = 10.1DD392 pKa = 3.27KK393 pKa = 11.34LGIKK397 pKa = 10.26GYY399 pKa = 10.86
Molecular weight: 44.9 kDa
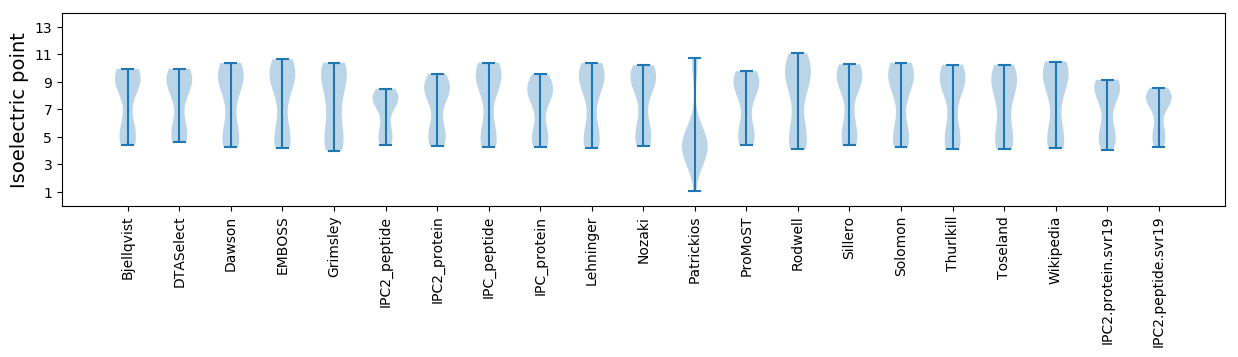
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0R5K4Q4|A0A0R5K4Q4_9VIRU Uncharacterized protein OS=Qinghai Lake virophage OX=1516115 GN=QLV_11 PE=4 SV=1
MM1 pKa = 7.6IEE3 pKa = 4.65AYY5 pKa = 9.86DD6 pKa = 3.71IKK8 pKa = 10.7KK9 pKa = 8.78TDD11 pKa = 4.12PLWKK15 pKa = 9.8YY16 pKa = 10.39SNPITAQNKK25 pKa = 7.18TYY27 pKa = 10.76SYY29 pKa = 10.17LGSDD33 pKa = 2.91ATLYY37 pKa = 10.92KK38 pKa = 9.55STRR41 pKa = 11.84EE42 pKa = 3.88GKK44 pKa = 10.37KK45 pKa = 10.86YY46 pKa = 9.4MVKK49 pKa = 10.28NPQGRR54 pKa = 11.84LIHH57 pKa = 6.5FGQLGYY63 pKa = 11.34EE64 pKa = 4.56DD65 pKa = 4.42FNKK68 pKa = 10.32HH69 pKa = 4.87KK70 pKa = 10.81SRR72 pKa = 11.84LRR74 pKa = 11.84RR75 pKa = 11.84RR76 pKa = 11.84AYY78 pKa = 10.68LNRR81 pKa = 11.84ATKK84 pKa = 8.79ITGDD88 pKa = 3.43WKK90 pKa = 10.54KK91 pKa = 11.12DD92 pKa = 3.09KK93 pKa = 10.96YY94 pKa = 11.28SPNNLSINILWW105 pKa = 3.71
MM1 pKa = 7.6IEE3 pKa = 4.65AYY5 pKa = 9.86DD6 pKa = 3.71IKK8 pKa = 10.7KK9 pKa = 8.78TDD11 pKa = 4.12PLWKK15 pKa = 9.8YY16 pKa = 10.39SNPITAQNKK25 pKa = 7.18TYY27 pKa = 10.76SYY29 pKa = 10.17LGSDD33 pKa = 2.91ATLYY37 pKa = 10.92KK38 pKa = 9.55STRR41 pKa = 11.84EE42 pKa = 3.88GKK44 pKa = 10.37KK45 pKa = 10.86YY46 pKa = 9.4MVKK49 pKa = 10.28NPQGRR54 pKa = 11.84LIHH57 pKa = 6.5FGQLGYY63 pKa = 11.34EE64 pKa = 4.56DD65 pKa = 4.42FNKK68 pKa = 10.32HH69 pKa = 4.87KK70 pKa = 10.81SRR72 pKa = 11.84LRR74 pKa = 11.84RR75 pKa = 11.84RR76 pKa = 11.84AYY78 pKa = 10.68LNRR81 pKa = 11.84ATKK84 pKa = 8.79ITGDD88 pKa = 3.43WKK90 pKa = 10.54KK91 pKa = 11.12DD92 pKa = 3.09KK93 pKa = 10.96YY94 pKa = 11.28SPNNLSINILWW105 pKa = 3.71
Molecular weight: 12.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6731 |
72 |
839 |
269.2 |
30.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.873 ± 0.814 | 1.307 ± 0.307 |
6.091 ± 0.435 | 6.522 ± 0.811 |
4.338 ± 0.378 | 6.983 ± 1.032 |
1.337 ± 0.261 | 7.488 ± 0.341 |
8.884 ± 1.264 | 8.439 ± 0.456 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.273 ± 0.241 | 6.864 ± 0.379 |
3.863 ± 0.369 | 3.684 ± 0.55 |
4.487 ± 0.532 | 6.091 ± 0.888 |
5.854 ± 0.532 | 4.962 ± 0.359 |
0.906 ± 0.183 | 4.754 ± 0.444 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
