
Candidatus Jidaibacter acanthamoeba
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rickettsiales; Candidatus Midichloriaceae; Candidatus Jidaibacter
Average proteome isoelectric point is 7.0
Get precalculated fractions of proteins
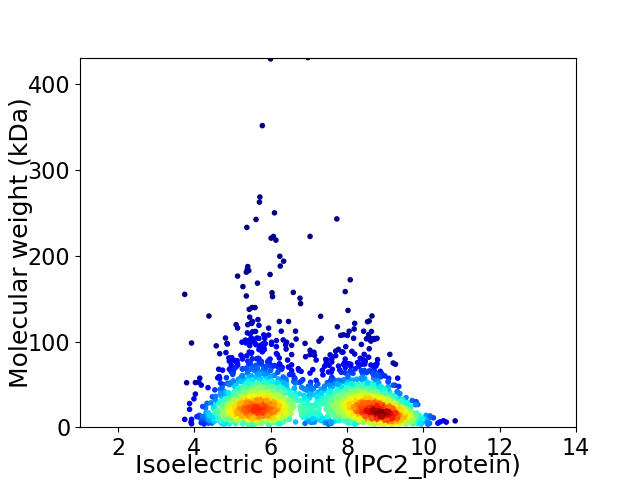
Virtual 2D-PAGE plot for 2204 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
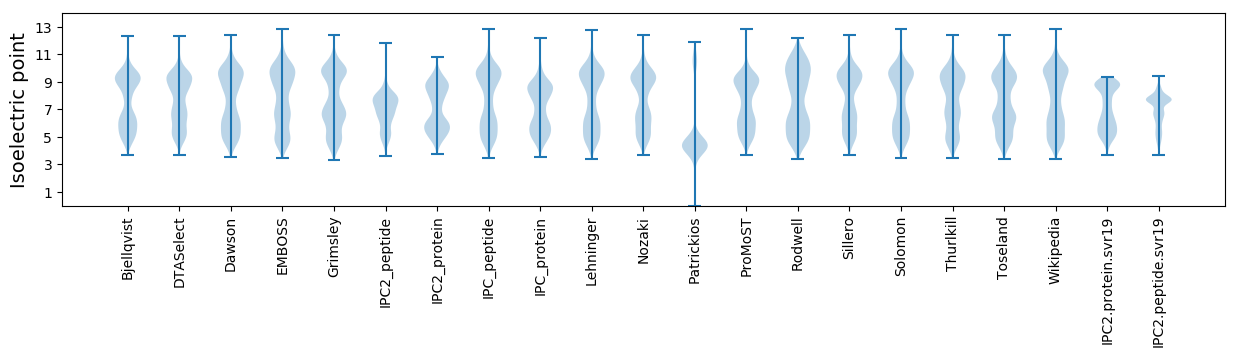
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C1R1F2|A0A0C1R1F2_9RICK Putative membrane protein insertion efficiency factor OS=Candidatus Jidaibacter acanthamoeba OX=86105 GN=NF27_BK00410 PE=3 SV=1
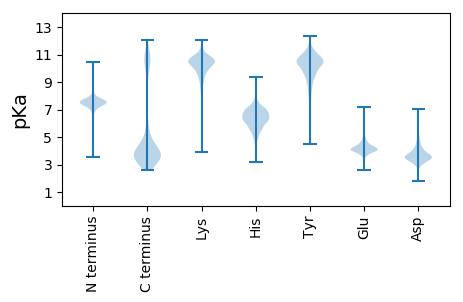
MM1 pKa = 7.4FNIGYY6 pKa = 10.52YY7 pKa = 10.88NMNSFKK13 pKa = 11.12NPFYY17 pKa = 10.82LSSLNGRR24 pKa = 11.84NGFAINGTTEE34 pKa = 3.83SMMEE38 pKa = 3.94SGFSVSRR45 pKa = 11.84AGDD48 pKa = 3.4INGDD52 pKa = 4.21GIADD56 pKa = 5.08LIIGAPGARR65 pKa = 11.84LKK67 pKa = 10.43MGASYY72 pKa = 11.32VVFGSQSPFSANTSLSDD89 pKa = 3.74LNGNNGFAIYY99 pKa = 10.46GRR101 pKa = 11.84TSLDD105 pKa = 3.38SLGVSVNGAGDD116 pKa = 3.52INGDD120 pKa = 4.06SIADD124 pKa = 4.69LIVSGAQTGAGYY136 pKa = 10.6VIFGSRR142 pKa = 11.84SPFPGNISLSDD153 pKa = 3.42INGKK157 pKa = 9.62NGFTISGLGNNYY169 pKa = 8.27WSKK172 pKa = 10.81PPVSGAGDD180 pKa = 3.4INADD184 pKa = 4.44GIADD188 pKa = 4.27MIIGDD193 pKa = 4.06SDD195 pKa = 3.52AGASYY200 pKa = 9.84TIFGSKK206 pKa = 10.52YY207 pKa = 9.37PFPANINVSEE217 pKa = 4.29LNGKK221 pKa = 9.24NGFTIYY227 pKa = 10.65GITSFDD233 pKa = 3.4SLGYY237 pKa = 9.3SVSGAGDD244 pKa = 3.29INADD248 pKa = 4.24GIADD252 pKa = 3.6IVIGAFTAGSNVGASYY268 pKa = 11.41VVFGSQSPFPTSINVYY284 pKa = 10.08SLNGKK289 pKa = 9.33NGFAINGIASLDD301 pKa = 3.64ALGSSVSGAGDD312 pKa = 3.32INGDD316 pKa = 4.23GIADD320 pKa = 5.08LIIGAPGASLKK331 pKa = 10.91AGASYY336 pKa = 10.97VIFGSQSTFPASINVSDD353 pKa = 4.88LNGKK357 pKa = 9.32NGFAINGITNPDD369 pKa = 3.42ALGSSVSGAGDD380 pKa = 3.32INGDD384 pKa = 4.15GIADD388 pKa = 4.72FIIGAPGAGSNVGASYY404 pKa = 11.41VVFGSQSFLPASINVSDD421 pKa = 4.05LNGRR425 pKa = 11.84NGFAIYY431 pKa = 9.64GAKK434 pKa = 10.09AYY436 pKa = 8.35EE437 pKa = 3.84NSGYY441 pKa = 10.16SVSGAGDD448 pKa = 3.46INDD451 pKa = 5.08DD452 pKa = 4.6GIADD456 pKa = 5.17LIIGAPEE463 pKa = 4.41AGSGTSYY470 pKa = 11.33VVFGMKK476 pKa = 8.82STTASLQISGSSYY489 pKa = 10.76SDD491 pKa = 3.07SLIYY495 pKa = 10.03EE496 pKa = 4.56DD497 pKa = 5.98KK498 pKa = 10.42EE499 pKa = 4.57TFLMDD504 pKa = 3.31TDD506 pKa = 4.14LTLKK510 pKa = 10.82LEE512 pKa = 4.31SEE514 pKa = 4.46II515 pKa = 4.81
MM1 pKa = 7.4FNIGYY6 pKa = 10.52YY7 pKa = 10.88NMNSFKK13 pKa = 11.12NPFYY17 pKa = 10.82LSSLNGRR24 pKa = 11.84NGFAINGTTEE34 pKa = 3.83SMMEE38 pKa = 3.94SGFSVSRR45 pKa = 11.84AGDD48 pKa = 3.4INGDD52 pKa = 4.21GIADD56 pKa = 5.08LIIGAPGARR65 pKa = 11.84LKK67 pKa = 10.43MGASYY72 pKa = 11.32VVFGSQSPFSANTSLSDD89 pKa = 3.74LNGNNGFAIYY99 pKa = 10.46GRR101 pKa = 11.84TSLDD105 pKa = 3.38SLGVSVNGAGDD116 pKa = 3.52INGDD120 pKa = 4.06SIADD124 pKa = 4.69LIVSGAQTGAGYY136 pKa = 10.6VIFGSRR142 pKa = 11.84SPFPGNISLSDD153 pKa = 3.42INGKK157 pKa = 9.62NGFTISGLGNNYY169 pKa = 8.27WSKK172 pKa = 10.81PPVSGAGDD180 pKa = 3.4INADD184 pKa = 4.44GIADD188 pKa = 4.27MIIGDD193 pKa = 4.06SDD195 pKa = 3.52AGASYY200 pKa = 9.84TIFGSKK206 pKa = 10.52YY207 pKa = 9.37PFPANINVSEE217 pKa = 4.29LNGKK221 pKa = 9.24NGFTIYY227 pKa = 10.65GITSFDD233 pKa = 3.4SLGYY237 pKa = 9.3SVSGAGDD244 pKa = 3.29INADD248 pKa = 4.24GIADD252 pKa = 3.6IVIGAFTAGSNVGASYY268 pKa = 11.41VVFGSQSPFPTSINVYY284 pKa = 10.08SLNGKK289 pKa = 9.33NGFAINGIASLDD301 pKa = 3.64ALGSSVSGAGDD312 pKa = 3.32INGDD316 pKa = 4.23GIADD320 pKa = 5.08LIIGAPGASLKK331 pKa = 10.91AGASYY336 pKa = 10.97VIFGSQSTFPASINVSDD353 pKa = 4.88LNGKK357 pKa = 9.32NGFAINGITNPDD369 pKa = 3.42ALGSSVSGAGDD380 pKa = 3.32INGDD384 pKa = 4.15GIADD388 pKa = 4.72FIIGAPGAGSNVGASYY404 pKa = 11.41VVFGSQSFLPASINVSDD421 pKa = 4.05LNGRR425 pKa = 11.84NGFAIYY431 pKa = 9.64GAKK434 pKa = 10.09AYY436 pKa = 8.35EE437 pKa = 3.84NSGYY441 pKa = 10.16SVSGAGDD448 pKa = 3.46INDD451 pKa = 5.08DD452 pKa = 4.6GIADD456 pKa = 5.17LIIGAPEE463 pKa = 4.41AGSGTSYY470 pKa = 11.33VVFGMKK476 pKa = 8.82STTASLQISGSSYY489 pKa = 10.76SDD491 pKa = 3.07SLIYY495 pKa = 10.03EE496 pKa = 4.56DD497 pKa = 5.98KK498 pKa = 10.42EE499 pKa = 4.57TFLMDD504 pKa = 3.31TDD506 pKa = 4.14LTLKK510 pKa = 10.82LEE512 pKa = 4.31SEE514 pKa = 4.46II515 pKa = 4.81
Molecular weight: 51.89 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C1QFZ1|A0A0C1QFZ1_9RICK Uncharacterized protein OS=Candidatus Jidaibacter acanthamoeba OX=86105 GN=NF27_HQ00070 PE=4 SV=1
MM1 pKa = 7.89SYY3 pKa = 10.46RR4 pKa = 11.84IPKK7 pKa = 8.73TLDD10 pKa = 2.88NPIRR14 pKa = 11.84CVGIPIDD21 pKa = 3.62TLMVFMVIWSALFLFDD37 pKa = 3.7QPIWGMVAGVIGANVFSRR55 pKa = 11.84YY56 pKa = 8.32RR57 pKa = 11.84SRR59 pKa = 11.84SITRR63 pKa = 11.84RR64 pKa = 11.84LVRR67 pKa = 11.84FIYY70 pKa = 9.47WYY72 pKa = 10.27LPCEE76 pKa = 4.1VNFIRR81 pKa = 11.84GVQGHH86 pKa = 4.57QRR88 pKa = 11.84KK89 pKa = 8.33MNMEE93 pKa = 4.32LKK95 pKa = 9.83KK96 pKa = 10.1WKK98 pKa = 10.27
MM1 pKa = 7.89SYY3 pKa = 10.46RR4 pKa = 11.84IPKK7 pKa = 8.73TLDD10 pKa = 2.88NPIRR14 pKa = 11.84CVGIPIDD21 pKa = 3.62TLMVFMVIWSALFLFDD37 pKa = 3.7QPIWGMVAGVIGANVFSRR55 pKa = 11.84YY56 pKa = 8.32RR57 pKa = 11.84SRR59 pKa = 11.84SITRR63 pKa = 11.84RR64 pKa = 11.84LVRR67 pKa = 11.84FIYY70 pKa = 9.47WYY72 pKa = 10.27LPCEE76 pKa = 4.1VNFIRR81 pKa = 11.84GVQGHH86 pKa = 4.57QRR88 pKa = 11.84KK89 pKa = 8.33MNMEE93 pKa = 4.32LKK95 pKa = 9.83KK96 pKa = 10.1WKK98 pKa = 10.27
Molecular weight: 11.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
678764 |
29 |
3780 |
308.0 |
34.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.489 ± 0.05 | 1.016 ± 0.016 |
4.84 ± 0.04 | 7.327 ± 0.071 |
4.443 ± 0.049 | 5.718 ± 0.055 |
1.869 ± 0.021 | 9.199 ± 0.056 |
8.592 ± 0.056 | 10.108 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.125 ± 0.024 | 6.332 ± 0.058 |
3.086 ± 0.033 | 3.238 ± 0.033 |
3.782 ± 0.047 | 6.871 ± 0.047 |
4.738 ± 0.041 | 5.392 ± 0.047 |
0.816 ± 0.016 | 4.014 ± 0.044 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
